Balancing accuracy, interpretability, and stability in machine-learning models: Live-weight prediction of Andean sheep from morphometric traits
DOI:
https://doi.org/10.17268/sci.agropecu.2025.037Palavras-chave:
biometrics, predictive models, mathematical models, young sheep, zoometricalResumo
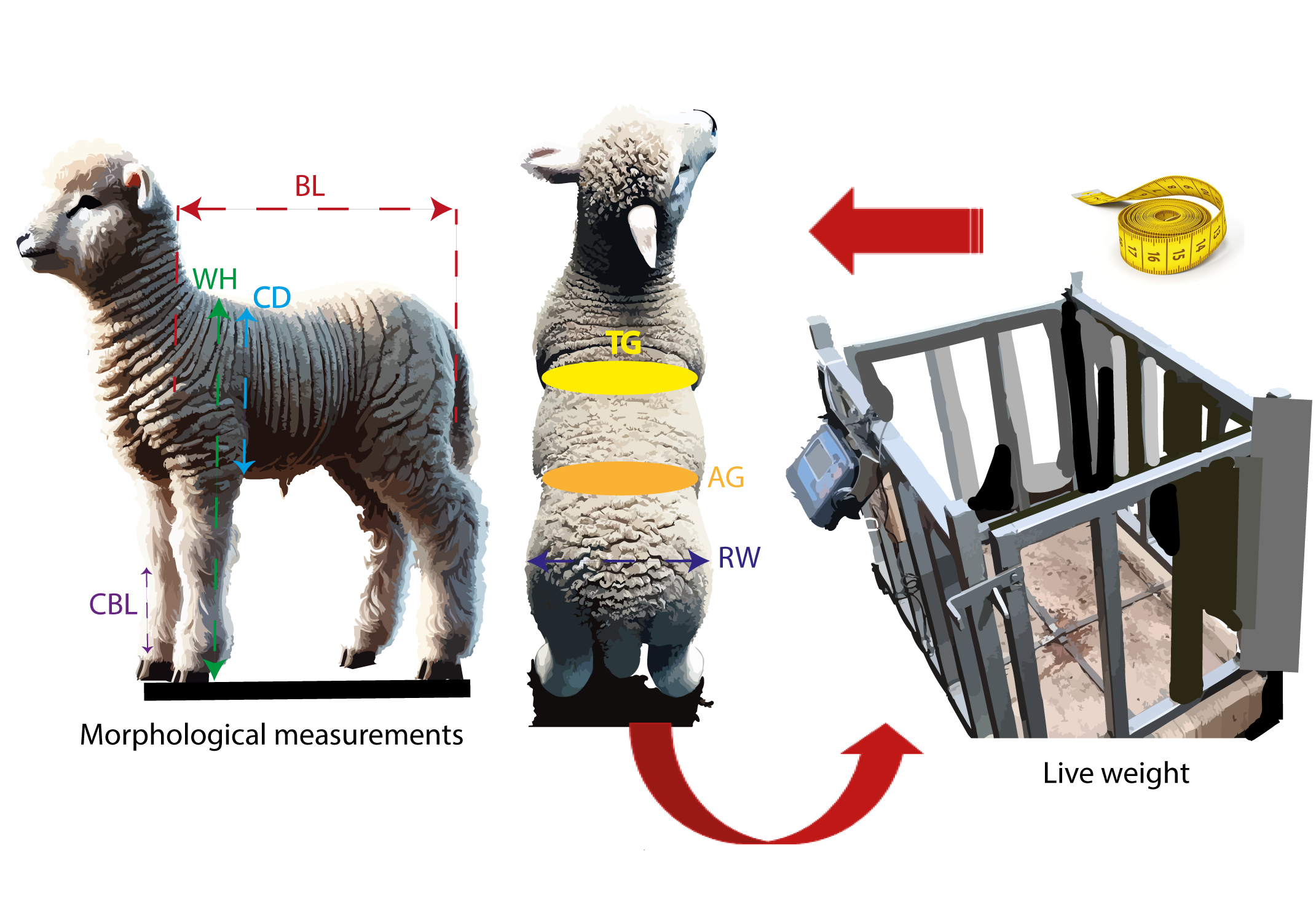
The objective of this research was to predict the live weight of Corriedale lambs using morphological measurements and machine learning algorithms. A total of 291 five-month-old lambs from the Corpacancha Production Unit of SAIS PACHACÚTEC SAC were used. These animals represented a homogeneous group in terms of age, sex, and genetics, as they belonged to the Corriedale breed and were offspring of "Category A" ewes. Morphological measurements recorded included Body Length (BL), Withers Height (WH), Thoracic Girth (TG), Rump Width (RW), Abdominal Girth (AG), Cannon Bone Length (CBL), Chest Depth (CD), and Live Weight (LW). The models evaluated were Multiple Linear Regression, Ridge Regression, Decision Trees, Random Forest, and XGBoost. The comparative analysis of the machine learning models identified ModG and Ridge as the most accurate and stable options, standing out for their low Mean Squared Error (MSE = 0.083) and Root Mean Squared Error (RMSE ≈ 0.287 – 0.288). Additionally, they exhibited the highest coefficients of determination (R2 = 0.89, RAdj2 = 0.88), indicating excellent predictive capability and data fit. Their low coefficient of variation (CV%) confirms their stability, establishing them as the best choices for applications where precision is paramount, such as predicting critical values in production processes and high-demand scientific studies. While XGBoost proved to be a robust alternative with an MSE of 0.119, an RMSE of 0.345, and a relative error of 2.22%. These findings confirm that prioritizing models that balance accuracy, interpretability, and stability enable faster, data-driven decision-making in Corriedale sheep production. Such an approach optimizes feed allocation, classifies lambs by market weight, and promptly detects growth deviations, thereby improving overall flock profitability.
Referências
Ahmed, H., Soliman, H., El-Sappagh, S., Abuhmed, T., & Elmogy, M. (2023). Early Detection of Alzheimer's Disease Based on Laplacian Re-Decomposition and XGBoosting. Computer Systems Science & Engineering, 46(3). https://doi.org/10.32604/csse.2023.036371
Arabameri, A., Tiefenbacher, J. P., Blaschke, T., Pradhan, B., & Tien Bui, D. (2020). Morphometric analysis for soil erosion susceptibility mapping using novel gis-based ensemble model. Remote Sensing, 12(5), 874. https://doi.org/10.3390/rs12050874
Bailey, D.W., Trotter, M.G., Tobin, C., & Thomas, M.G. (2021). Opportunities to apply precision livestock management on rangelands. Frontiers in Sustainable Food Systems, 5, 611915. https://doi.org/10.3389/fsufs.2021.611915
Cam, M. A., Olfaz, M., & Soydan, E. (2010). Body measurements reflect body weights and carcass yields in Karayaka sheep. Asian Journal of Animal and Veterinary Advances, 5(2), 120-127. https://doi.org/10.3923/ajava.2010.120.127
Carhuas, J. N., Capcha, K. B., Garcia-Olarte, E., & Eulogio, C. Q. (2024). Production performance of rejected newborn lambs fed with different concentrations of whey in Perú. Revista De Ciências Agroveterinárias, 23(2), 231–239. https://doi.org/10.5965/223811712322024231
Choque, C. J. B. (2024). Mathematical models of chlorine demand in river waters: a systematic review. Tecnia, 34(1), 26-41. https://doi.org/10.21754/tecnia.v34i1.1635
Contreras, J. P., Cordero, A., Rojas, Y., Carhuas, J., Curasma, J., Mayhua, P., & Salazar, K. (2024). Prediction models for live body weight and body compactness of Criollo sheep in Huancavelica Region, Peru. The Indian Journal of Animal Sciences, 94(7), 637-641. https://doi.org/10.56093/ijans.v94i7.148186
Courtenay, L. A., Yravedra, J., Huguet, R., Aramendi, J., Maté-González, M. Á., González-Aguilera, D., & Arriaza, M. C. (2019). Combining machine learning algorithms and geometric morphometrics: a study of carnivore tooth marks. Palaeogeography, Palaeoclimatology, Palaeoecology, 522, 28-39. https://doi.org/10.1016/j.palaeo.2019.03.007
Dang, C., Choi, T., Lee, S., Lee, S., Alam, M., Park, M., ... & Hoang, D. (2022). Machine learning-based live weight estimation for hanwoo cow. Sustainability, 14(19), 12661. https://doi.org/10.3390/su141912661
Ergin, M., & Koşkan, Ö. (2025). Estimating body weight in Sujiang pigs using artificial neural network, nearest neighbor, and CART algorithms: a comparative study using morphological measurements. Tropical Animal Health and Production, 57(1), 17. https://doi.org/10.1007/s11250-024-04258-7
Frizzarin, M., Gormley, I. C., Berry, D. P., Murphy, T. B., Casa, A., Lynch, A., & McParland, S. (2021). Predicting cow milk quality traits from routinely available milk spectra using statistical machine learning methods. Journal of Dairy Science, 104(7), 7438-7447. https://doi.org/10.3168/jds.2020-19576
García-Medina, A., & Aguayo-Moreno, E. (2024). LSTM–GARCH hybrid model for the prediction of volatility in cryptocurrency portfolios. Computational Economics, 63(4), 1511-1542. https://doi.org/10.1007/s10614-023-10373-8
Gomes, R.A., Monteiro, G.R., Assis, G.J., Busato, K.C., Ladeira, M.M., & Chizzotti, M.L. (2016). Technical note: Estimating body weight and body composition of beef cattle trough digital image analysis. Journal of Animal Science, 94, 5414–5422. https://doi.org/10.2527/jas.2016-0797
Gonçalves, M. A., Castro, M. S. M., Carrara, E. R., Raineri, C., Rennó, L. N., & Schultz, E. B. (2025). Prediction of Weight and Body Condition Score of Dairy Goats Using Random Forest Algorithm and Digital Imaging Data. Animals, 15(10), 1449. https://doi.org/10.3390/ani15101449
Hu, J., & Szymczak, S. (2023). A review on longitudinal data analysis with random forest. Briefings in Bioinformatics, 24(2), bbad002.
Jarupunphol, P., Buathong, W., Kuptabut, S., & Sudjarid, W. (2025). Assessing decision tree, random forest, and XGBoost models for human capital readiness predictions in low-income areas. Multidisciplinary Science Journal, 7(6), 2025296-2025296. https://doi.org/10.31893/multiscience.2025296
Jurkovich, V., Hejel, P., & Kovács, L. (2024). A review of the effects of stress on dairy cattle behaviour. Animals, 14(14), 2038. https://doi.org/10.3390/ani14142038
Karna, D. K., Mishra, C., Dash, S. K., Acharya, A. P., Panda, S., & Chinnareddyvari, C. S. (2024). Exploring body morphometry and weight prediction in Ganjam goats in India through principal component analysis. Tropical Animal Health and Production, 56(8), 298. https://doi.org/10.1007/s11250-024-04114-8
Kumar, R., Sharma, D., Dua, A., & Jung, K. H. (2023). A review of different prediction methods for reversible data hiding. Journal of Information Security and Applications, 78, 103572. https://doi.org/10.1016/j.jisa.2023.103572
Kozaklı, Ö., Ceyhan, A., & Noyan, M. (2024). Comparison of machine learning algorithms and multiple linear regression for live weight estimation of Akkaraman lambs. Tropical Animal Health and Production, 56(7), 250. https://doi.org/10.1007/s11250-024-04049-0
Lee, C. S., Cheang, P. Y. S., & Moslehpour, M. (2022). Predictive analytics in business analytics: decision tree. Advances in Decision Sciences, 26(1), 1-29. https://doi.org/10.47654/v26y2022i1p1-29
Liang, Z., Cai, L., Wang, S., & Wang, Q. (2025). K-fold cross-validation based frequentist model averaging for linear models with nonignorable missing responses. Statistics and Computing, 35(1), 18. https://doi.org/10.1007/s11222-024-10554-x
Lipovetsky, S. (2021). Modified ridge and other regularization criteria: A brief review on meaningful regression models. Model Assisted Statistics and Applications, 16(3), 225-227. https://doi.org/10.3233/MAS-210536
Long, K., Guo, D., Deng, L., Shen, H., Zhou, F., & Yang, Y. (2025). Cross-Combination Analyses of Random Forest Feature Selection and Decision Tree Model for Predicting Intraoperative Hypothermia in Total Joint Arthroplasty. The Journal of Arthroplasty, 40(1), 61-69. https://doi.org/10.1016/j.arth.2024.07.007
Mahmud, M. A., Shaba, P., Abdulsalam, W., Yisa, H. Y., Gana, J., Ndagi, S., & Ndagimba, R. (2014). Live body weight estimation using cannon bone length and other body linear measurements in Nigerian breeds of sheep. Journal of Advanced Veterinary and Animal Research, 1(4), 169-176. https://doi.org/10.5455/javar.2014.a29
Martins, B. M., Mendes, A. L., Silva, L. F., Moreira, T. R., Costa, J. H., Rotta, P. P., Chizzotti, M. L., & Marcondes, M. I. (2020). Estimating body weight, body condition score, and type traits in dairy cows using three dimensional cameras and manual body measurements. Livestock Science, 236, 104054. https://doi.org/10.1016/j.livsci.2020.104054
Mokri, M., Safari, M., Kaviani, S., Juneau, D., Cohalan, C., Archambault, L., & Carrier, J. F. (2025). Deep learning-based prediction of later 13N-ammonia myocardial PET image frames from initial frames. Biomedical Signal Processing and Control, 100, 106865.
Ninahuanca Carhuas, J., Cerna, L, A., Unchupaico Payano, I, Garcia-Olarte, E., Mauricio-Ramos, Y., Quispe Eulogio, C., & Hadi Mohamed, Mohamed M. (2025). Counting sheep: human experience vs. Yolo algorithm with drone to determine population. Veterinary Integrative Sciences, 23(2), 1-9. https://doi.org/10.12982/VIS.2025.032
Ozen, H., Ozen, D., Kocakaya, A., & Ozbeyaz, C. (2024). Shrinkage and tree-based regression methods for the prediction of the live weight of Akkaraman sheep using morphological traits. Tropical Animal Health and Production, 56(8), 346. https://doi.org/10.1007/s11250-024-04187-5
Pannier, L., Tarr, G., Pleasants, T., Ball, A., McGilchrist, P., Gardner, G. E., & Pethick, D. W. (2025). The construction of a sheepmeat eating quality prediction model for Australian lamb. Meat science, 220, 109711. https://doi.org/10.1016/j.meatsci.2024.109711
Peña-Avelino, L. Y., Alva-Pérez, J., Ceballos-Olvera, I., Hernández-Contreras, S., & Álvarez-Fuentes, G. (2021). Evaluación de diferentes fórmulas zoométricas para la estimación de peso vivo en cabras criollas de Tamaulipas, México. Producción Vegetal, 532. https://doi.org/10.12706/itea.2021.007
Qin, Q., Zhang, C. Y., Liu, Z. C., Wang, Y. C., Kong, D. Q., Zhao, D., ... & Liu, Z. H. (2024). Estimation of the genetic parameters of sheep growth traits based on machine vision acquisition. Animal, 18(7), 101196. https://doi.org/10.1016/j.animal.2024.101196
Salamanca-Carreño, A., Parés-Casanova, P. M., Vélez-Terranova, M., Martínez-Correal, G., & Rangel-Pachón, D. E. (2024). Early Cannon Development in Females of the “Sanmartinero” Creole Bovine Breed. Animal, 14(4), 527. https://doi.org/10.3390/ani14040527
Samuel, A.L. Algunos estudios en aprendizaje automático utilizando el juego de damas. Revista de investigación y desarrollo de IBM. 2000, 44206–226.
Shen, Y., Wu, S., Wang, Y., Wang, J., & Yang, Z. (2025). Interpretable model for rockburst intensity prediction based on Shapley values-based Optuna-random forest. Underground Space, 21, 198-214. https://doi.org/10.1016/j.undsp.2024.09.002
Stewart, W. C., Scasta, J. D., Maierle, C., Ates, S., Burke, J. M., & Campbell, B. J. (2025). Vegetation management utilizing sheep grazing within utility-scale solar: Agro-ecological insights and existing knowledge gaps in the United States. Small Ruminant Research, 243, 107439. https://doi.org/10.1016/j.smallrumres.2025.107439
Vlaicu, P. A., Gras, M. A., Untea, A. E., Lefter, N. A., & Rotar, M. C. (2024). Advancing Livestock Technology: Intelligent Systemization for Enhanced Productivity, Welfare, and Sustainability. AgriEngineering, 6(2), 1479-1496. https://doi.org/10.3390/agriengineering6020084
Wang, Z., Shadpour, S., Chan, E., Rotondo, V., Wood, K. M., & Tulpan, D. (2021) Applications of machine learning for livestock body weight prediction from digital images. Journal of Animal Science, 99(2), skab022. https://doi.org/10.1093/jas/skab022
Downloads
Arquivos adicionais
Publicado
Como Citar
Edição
Seção
Licença
Copyright (c) 2025 Scientia Agropecuaria

Este trabalho está licenciado sob uma licença Creative Commons Attribution-NonCommercial 4.0 International License.
Los autores que publican en esta revista aceptan los siguientes términos:
a. Los autores conservan los derechos de autor y conceden a la revista el derecho publicación, simultáneamente licenciada bajo una licencia de Creative Commons que permite a otros compartir el trabajo, pero citando la publicación inicial en esta revista.
b. Los autores pueden celebrar acuerdos contractuales adicionales separados para la distribución no exclusiva de la versión publicada de la obra de la revista (por ejemplo, publicarla en un repositorio institucional o publicarla en un libro), pero citando la publicación inicial en esta revista.
c. Se permite y anima a los autores a publicar su trabajo en línea (por ejemplo, en repositorios institucionales o en su sitio web) antes y durante el proceso de presentación, ya que puede conducir a intercambios productivos, así como una mayor citación del trabajo publicado (ver efecto del acceso abierto).




