Genetic diversity of antagonistic bacterial isolates obtained from Theobroma cacao L. to control Fusarium oxysporum f. sp. cubense race 1
DOI:
https://doi.org/10.17268/sci.agropecu.2025.047Keywords:
Indole-3-acetic acid, gibberellic acid, salicylic acid, cell extracts, ChiA gene, Musa acuminataAbstract
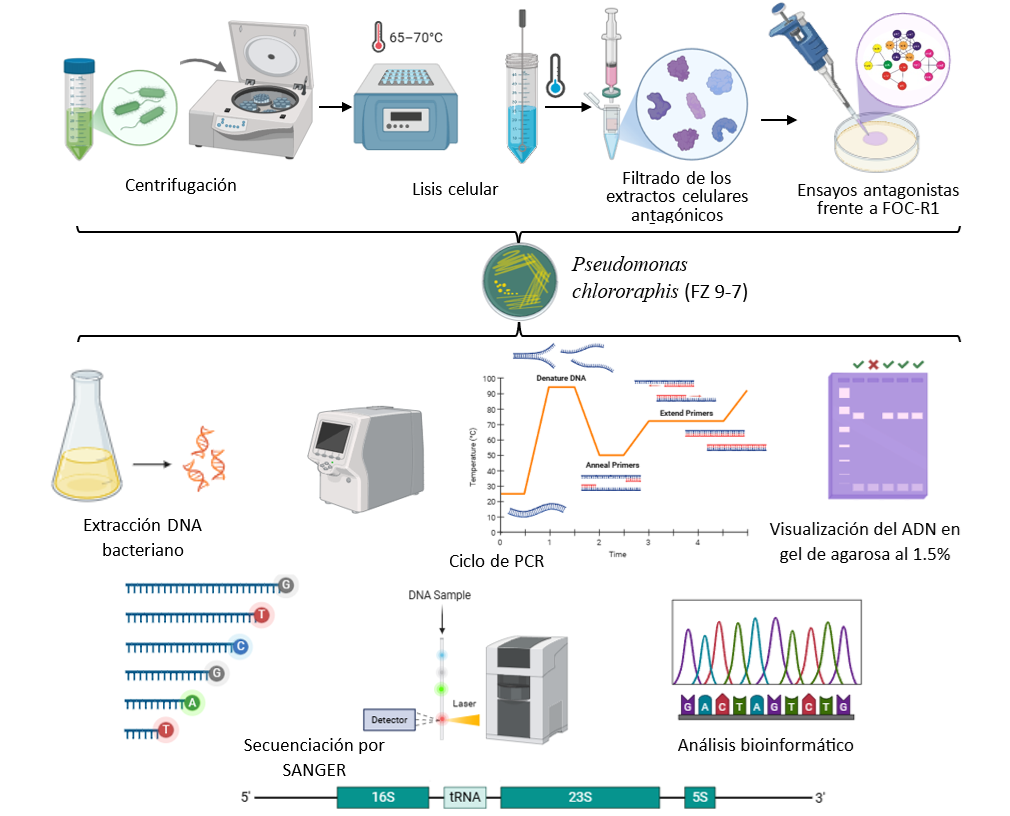
The banana (Musa AAA) is affected by Fusarium oxysporum f. sp. cubense, which causes discoloration in the xylem duct, leading to terminal wilting. The use of plant growth promoting rhizobacteria (PGPR) as a biological control produces different antagonistic compounds and inhibits the growth of various phytopathogens. The objective of the study was based on the molecular identification of rhizobacteria that produce phytohormones with biocontrol activity against Foc-R1. The presence of the 225 bp ChiA gene was observed in PGPR. Phylogenetic analysis of 16S rRNA by sequencing and ERIC-PCR showed genetic variability with the formation of four subgroups. Molecular identification by sequencing the 16S rRNA gene defined the genera as Klebsiella, Enterobacter, and Pseudomonas. There is variation in the biosynthesis of the phytohormones AIA, AG, and AS in strains MH-18, W-417, and FZ 9-7 at 72 h. The identification of Foc-R1 by PCR shows an amplicon of 350 bp. Antagonistic assays of bacterial supernatants from strain FZ 9-7 show 71% mycelial inhibition of Foc-R1 and a decrease in spore production of 2.5X106 spores mL-1. The results provide information on the genetic relationships of PGPRs through the production of secondary metabolites such as proteases, catalases, chitinases, and siderophores, as well as morphological and molecular analysis for the identification of Foc-R1 and its interaction with antagonistic extracts in inhibiting the growth of diseases in bananas and cocoa.
References
Aguilar-Hawod, K. G. I., de la Cueva, F. M., & Cumagun, C. J. R. (2019). Genetic diversity of Fusarium oxysporum f. sp. cubense causing Panama wilt of banana in the Philippines. Pathogens, 9(1), 32. https://doi.org/10.3390/pathogens9010032
Arrebola, E., Aprile, F. R., Calderón, C. E., De Vicente, A., & Cazorla, F. M. (2022). Insecticidal features displayed by the beneficial rhizobacterium Pseudomonas chlororaphis PCL1606. International Microbiology, 25(4), 679-689. https://doi.org/10.1007/s10123-022-00253-w
Asril, M., & Supriyadi, D. (2024). Characterization of Extracellular Chitinase from Bacillus cereus SAHA 12.13 and Its Potency as a Biocontrol of Curvularia affinis. Journal of Multidisciplinary Applied Natural Science, 4(1), 165-175. https://doi.org/10.47352/jmans.2774-3047.203
Auhing Arcos, J. A., Cedeño Moreira, Á. V., Saucedo Aguiar, S., Vera Benites, L. F., Macías Holguín, C. J., & Martínez, H. F. C. (2021). Biodiversidad de ecotipos y rangos de agresividad de Moniliophthora perniciosa, en Theobroma cacao L. nacional del Litoral Ecuatoriano. Scientia Agropecuaria, 12(4), 599-609. http://dx.doi.org/10.17268/sci.agropecu.2021.064
Baldy-Chudzik, K., & Stosik, M. (2005). Specific genomic fingerprints of Escherichia coli strains with repetitive sequences and PCR as an effective tool for monitoring freshwater environments. Polish Journal of Environmental Studies, 14(5).
Beale, M. H., Bearder, J. R., Down, G. H., Hutchison, M., MacMillan, J., & Phinney, B. O. (1982). The biosynthesis of kaurenolide diterpenoids by Gibberella fujikuroi. Phytochemistry, 21(6), 1279-1287. https://doi.org/10.1016/0031-9422(82)80126-X
Bharucha, U., Patel, K., & Trivedi, U. B. (2013). Optimization of indole acetic acid production by Pseudomonas putida UB1 and its effect as plant growth-promoting rhizobacteria on mustard (Brassica nigra). Agricultural research, 2, 215-221. https://doi.org/10.1007/s40003-013-0065-7.
Canchignia-Martínez, H. F., Macías-Holguin, C. J., Tapia-Quintana, D. N., Manzo-Campos, T., Saltos-Avilés, J. D., & Vera-Benites, L. F. (2025a). Bacterias Productoras de Ácido Indol-3-acético y Solubilizadoras de Fósforo y Potasio como Promotoras de Crecimiento en Oryza sativa L. Revista Terra Latinoamericana, 43. https://doi.org/10.28940/terralatinoamericana.v43i.1969
Canchignia-Martínez, H. F., Macías-Holguín, C. J., Zurita Segovia, R. D., Vera-Benites, L. F., Ortiz-Almea, H. G., & Tapia-Quintana, D. N. (2025b). Rhizobacteria and Cell Extracts with Antagonistic Potential on Phytophthora palmivora in Theobroma cacao CCN-51. Revista Terra Latinoamericana, 43. https://doi.org/10.28940/terralatinoamericana.v43i.2009
Canchignia-Martínez, H. F., Vera-Benites, L. F., Tapia-Quintana, D. N., Cedeño-Moreira, Á. V., García-Intriago, E., & Macías-Holguín, C. J. (2024c). Caracterización Bioquímica de Rizobacterias Endófitas con Actividad Biocontroladora Contra Phytophthora palmivora y Lasiodiplodia theobromae. Revista Terra Latinoamericana, 42. https://doi.org/10.28940/terra.v42i0.1807
Cedeño Moreira, Á. V., Romero Meza, R. F., Auhing Arcos, J. A., Mendoza León, A. F., Abasolo Pacheco, F., & Canchignia Martínez, H. F. (2020). Caracterización de Phytophthora spp. y aplicación de rizobacterias con potencial en biocontrol de la enfermedad de la mazorca negra en Theobroma cacao variedad CCN-51. Scientia Agropecuaria, 11(4), 503-512. https://dx.doi.org/10.17268/sci.agropecu.2020.04.05
Chávez-Arteaga, K., Guato-Molina, J., Peñafiel-Jaramillo, M., Mestanza-Uquillas, C., & Canchignia-Martínez, H. F. (2018). Bacterias fluorescentes productoras de metabolitos antagónicos de cultivares nativos de Musa sp. y su diversidad filogenética al gen ARNr 16S. Ciencia y Tecnología, 11, 17-29.
Chávez-Arteaga, K. T., Guato Molina, J. J., Rodríguez Acosta, J. L., Cedeño Moreira, Ángel V., Romero Meza, R. F., Canchignia Martínez, H. F. (2020). Rizobacterias con potencial antagonista in vitro a Mycosphaerella fijiensis Morelet. Ciencia y Tecnología, 13(2), 9–16. https://doi.org/10.18779/cyt.v13i2.387
Chen, Q., Qi, P., Xu, R., Tambong, J. T., Djama, Z. R., & Li, W. (2011). Comparison of three typing methods for evaluating the diversity of Pseudomonas fluorescens in the rhizosphere. Journal of Plant Sciences, 6(2), 52. https://dx.doi.org/10.3923/jps.2011.52.65
Cook, R. J. (1993). Making greater use of introduced microorganisms for biological control of plant pathogens. Annual review of phytopathology, 31(1), 53-80. https://doi.org/10.1146/annurev.py.31.090193.000413
Crespo Ávila, J. A., Carranza Cárdenas, C. C., Cedeño Moreira, A. V., Vera Benites, L. F., & Chevez Villanueva, M. S. (2024). Actividad antagonista de PGPR en nematodo fitoparásito Pratylenchus spp. en Musa paradisiaca (Musa acuminata× M. balbisiana) vc cavendish. Alfa Revista de Investigación en Ciencias Agronómicas y Veterinaria, 8(24), 717-728. https://doi.org/10.33996/revistaalfa.v8i24.297
Crespo-Clas, Á. M., Cedeño-Moreira, Á. V., Canchignia-Martínez, H. F., & Garcés-Fiallos, F. R. (2024). Rhizobacterial consortium differently affects black leaf spot, physiological, morpho-logical, and productive components in two generations of banana plants. Rhizosphere, 31, 100932. https://doi.org/10.1016/j.rhisph.2024.100932
da Silva, C. B., Dos Santos, H. R. M., Marbach, P. A. S., de Souza, J. T., Cruz-Magalhães, V., Argôlo-Filho, R. C., & Loguercio, L. L. (2019). First-tier detection of intragenomic 16S rRNA gene variation in culturable endophytic bacteria from cacao seeds. PeerJ, 7, e7452. https://doi.org/10.7717/peerj.7452
Dita, M. A., Waalwijk, C., Buddenhagen, I. W., Souza Jr, M. T., & Kema, G. H. J. (2010). A molecular diagnostic for tropical race 4 of the banana fusarium wilt pathogen. Plant pathology, 59(2), 348-357. https://doi.org/10.1111/j.1365-3059.2009.02221.x
Djeugap, J. F., Abireche, H. U., Donfack, C. P., Sonkoue, A. M., Ndogho, A., & Nouteka, J. N. (2023). Cultural characterization of five isolates of Fusarium oxysporum f. sp. cubense (banana fusarium wilt) and antifungal activity of plant extracts. Pakistan Journal of Phytopathology, 35(1), 43-53. https://doi.org/10.33866/phytopathol.035.01.0844
Drancourt, M., Bollet, C., & Raoult, D. (2000). 16 S ribosomal DNA sequence analysis of a large collection of enviromental and clinical unidentiable bacterial isolates. Journal of Clinical Microbiol, 38: 362-363. https://doi.org/10.1128/jcm.38.10.3623-3630.2000
El-Sapagh, S., Allam, N. G., El-Sayed, M. N. E. D., El-Hefnawy, A. A., Korbecka-Glinka, G., & Shala, A. Y. (2023). Effects of Silybum marianum L. seed extracts on Multi Drug Resistant (MDR) Bacteria. Molecules, 29(1), 64. https://doi.org/10.3390/molecules29010064
Fan, M., Tan, S., Wang, W., & Zhang, X. (2024). Improvement in Salt Tolerance Ability of Pseudomonas putida KT2440. Biology, 13(6), 404. https://doi.org/10.3390/biology13060404
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. evolution, 39(4), 783-791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Flury, P., Vesga, P., Péchy-Tarr, M., Aellen, N., Dennert, F., et al. (2017). Antimicrobial and insecticidal: cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Frontiers in microbiology, 8, 100. https://doi.org/10.3389/fmicb.2017.00100.
Gang, S., Sharma, S., Saraf, M., Buck, M., & Schumacher, J. (2019). Analysis of indole-3-acetic acid (IAA) production in Klebsiella by LC-MS/MS and the Salkowski method. Bio-protocol, 9(9), e3230-e3230. https://doi.org/10.21769/BIOPROTOC.3230
Getahun, A., Kiros, S., Muleta, D., & Assefa, F. (2020). Genetic and metabolic diversities of rhizobacteria isolated from degraded soil of Ethiopia. Heliyon, 6(12). https://doi.org/10.1016/j.heliyon.2020.e05697
Gordon, S. A., & Weber, R. P. (1951). Colorimetric estimation of indoleacetic acid. Plant physiology, 26(1), https://doi.org/192. 10.1104/pp.26.1.192
Guato-Molina, J. J., Auhing-Arcos, J. A., Crespo-Ávila, J. A., Esmeraldas-García, G. A., Mendoza-León, A. F., & Canchignia-Martínez, H. F. (2019). Bacterias promotoras del crecimiento en plantas con potencial agente biocontrolador a Fusarium oxysporum f. sp. Lycopersici, y Moniliophthora roreri. Scientia Agropecuaria, 10(3), 393-402. http://dx.doi.org/10.17268/sci.agropecu.2019.03.10
Gupta, N., Balomajumder, C., & Agarwal, V. K. (2010). Enzymatic mechanism and biochemistry for cyanide degradation: a review. Journal of hazardous materials, 176(1-3), 1-13. https://doi.org/10.1016/j.jhazmat.2009.11.038
Gurdaswani, V., Ghag, S. B., & Ganapathi, T. R. (2020). FocSge1 in Fusarium oxysporum f. sp. cubense race 1 is essential for full virulence. BMC microbiology, 20, 1-15. https://doi.org/10.4067/S0717-34582008000500005
Gusmiaty, Restu, M., Bachtiar, B., & Larekeng, S. H. (2019). Gibberellin and IAA production by rhizobacteria from various private forest. IOP Conference Series: Earth and Environmental Science, 270(1), 012018. https://doi.org/10.1088/1755-1315/270/1/012018.
Hernández García, M., Morgante, V., Avila Perez, M., Villalobos Biaggini, P., Miralles Noé, P., González Vergara, M., & Seeger Pfeiffer, M. (2008). Novel s-triazine-degrading bacteria isolated from agricultural soils of central Chile for herbicide bioremediation. Electronic Journal of Biotechnology, 11(5), 5-6. http://dx.doi.org/10.4067/S0717-34582008000500005
Hernández, A., Rives, N., Caballero, A., Hernández, A. N., & Heydrich, M. (2004). Caracterización de rizobacterias asociadas al cultivo del maíz en la producción de metabolitos del tipo AIA, sideróforos y ácido salicílico. Revista Colombiana de biotecnología, 6(1), 6-13.
Holbrook, A. A., Edge, W. J. W., & Bailey, F. (1961). Spectrophotometric method for determination of gibberellic acid. https://doi.org/10.1021/ba-1961-0028.ch018
Izquierdo-García, L. F., Carmona, S. L., Zuluaga, P., Rodríguez, G., Dita, M., Betancourt, M., & Soto-Suárez, M. (2021). Efficacy of disinfectants against Fusarium oxysporum f. sp. cubense tropical race 4 isolated from La Guajira, Colombia. Journal of Fungi, 7(4), 297. https://doi.org/10.3390/jof7040297
Jaroszuk-Ściseł, J., Tyśkiewicz, R., Nowak, A., Ozimek, E., Majewska, M., Hanaka, A., ... & Janusz, G. (2019). Phytohormones (auxin, gibberellin) and ACC deaminase in vitro synthesized by the mycoparasitic Trichoderma DEMTkZ3A0 strain and changes in the level of auxin and plant resistance markers in wheat seedlings inoculated with this strain conidia. International Journal of Molecular Sciences, 20(19), 4923. https://doi.org/10.3390/ijms20194923
Kalimuthu, R., Suresh, P., Varatharaju, G., Balasubramanian, N., Rajasekaran, K. M., & Shanmugaiah, V. (2019). Isolation and characterization of Indole acetic acid [IAA] producing tomato Rhizobacterium pseudomonas sp VSMKU4050 and its potential for plant growth promotion. International Journal of Current Microbiology and Applied Sciences, 8(06), 443-455. https://doi.org/10.20546/ijcmas.2019.806.050
Kang, S. M., Adhikari, A., Lee, K. E., & Park, Y. G. (2019). Gibberellin producing rhizobacteria Pseudomonas koreensis MU2 enhance growth of lettuce (Lactuca sativa) and Chinese cabbage (Brassica rapa, chinensis). Journal of Microbiology, Biotechnology & Food Sciences, 9(2). https://doi.org/10.15414/jmbfs.2019.9.2.166-170
Kesaulya, H., Zakaria, B., & Syaiful, S. A. (2015). Isolation and physiological characterization of PGPR from potato plant rhizosphere in medium land of Buru Island. Procedia Food Science, 3, 190-199. https://doi.org/10.1016/j.profoo.2015.01.021
Keswani, C., Singh, S. P., García‐Estrada, C., Mezaache‐Aichour, S., Glare, T. R., Borriss, R., ... & Sansinenea, E. (2022). Biosynthesis and beneficial effects of microbial gibberellins on crops for sustainable agriculture. Journal of applied microbiology, 132(3), 1597-1615. https://doi.org/10.1111/JAM.15348
King, E. O., Ward, M. K., & Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. The Journal of laboratory and clinical medicine, 44(2), 301-307. https://doi.org/10.5555/URI:PII:002221435490222X
Kochar, M., Upadhyay, A., & Srivastava, S. (2011). Indole-3-acetic acid biosynthesis in the biocontrol strain Pseudomonas fluorescens Psd and plant growth regulation by hormone overexpression. Research in microbiology, 162(4), 426-435. https://doi.org/10.1016/j.resmic.2011.03.006
Lee, Y. S., & Kim, K. Y. (2015). Statistical optimization of medium components for chitinase production by Pseudomonas fluorescens strain HN1205: role of chitinase on egg hatching inhibition of root-knot nematode. Biotechnology & Biotechnological Equipment, 29(3), 470-478. https://doi.org/10.1080/13102818.2015.1010702
Lenin, G., & Jayanthi, M. (2012). Indole acetic acid, gibberellic acid and siderophore production by PGPR isolates from rhizospheric soils of Catharanthus roseus. International Journal of Pharmaceutical Biological Archive, 3(4), 933-938.
Li, M. H., Yu, X. T., Wang, H. F., Zhou, J. N., Xi, P. G., & Jiang, Z. D. (2012). Rapid detection and identification of Fusarium oxysporum f. sp. cubense race 1 and race 4. Scientia Agricultura Sinica, 45(19), 3971-3979. http://10.3864/j.issn.0578-1752.2012.19.008
López-Zapata, S. P., & Castaño-Zapata, J. (2019). Integrated management of Panama disease Fusarium oxysporum Schlechtend.: Fr. f. sp. cubense (EF SM.) WC Snyder & HN Hansen: a review. Revista UDCA Actualidad & Divulgación Científica, 22(2). https://doi.org/10.31910/rudca.v22.n2.2019.1240
Louws, F. J., Fulbright, D. W., Stephens, C. T., & De Bruijn, F. J. (1994). Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Applied and environmental microbiology, 60(7), 2286-2295. https://doi.org/10.1128/aem.60.7.2286-2295.1994
Macías-Holguín, C. J., Canchignia-Martínez, H. F., Delgado-Basurto, V. D., Paucar-Nieto, F. P., Arellano-Ibarra, K. V., & Cedeño-Moreira, Á. V. (2023a). Efectos de la co-inoculación de Bioformulados (PGPR´ s) sobre el porcentaje de germinación y promover el crecimiento en plántula de papaya (Carica papaya L.). Manglar, 20(2), 149-155. http://dx.doi.org/10.57188/manglar.2023.017
Macías Holguín, C. J., Valarezo Padilla, F. C., Tapia Quintana, D. N., Canchignia Martínez, H. F., Cedeño Moreira, Ángel V., & García Intriago, E. (2023b). Efecto de bioformulados bacterianos como controladores de Radopholus similis y potenciadores del desarrollo de plántulas de banano (Musa acuminata) cultivar Williams. Ciencia y Tecnología, 16(2), 9–16. https://doi.org/10.18779/cyt.v16i2.705
Magdama, F., Monserrate-Maggi, L., Serrano, L., Sosa, D., Geiser, D. M., & Jiménez-Gasco, M. D. M. (2019a). Comparative analysis uncovers the limitations of current molecular detection methods for Fusarium oxysporum f. sp. cubense race 4 strains. PLoS One, 14(9), e0222727. https://doi.org/10.1371/JOURNAL.PONE.0222727
Magdama, F., Monserrate-Maggi, L., Serrano, L., García Onofre, J., & Jiménez-Gasco, M. D. M. (2020b). Genetic Diversity of Fusarium oxysporum f. sp. cubense, the Fusarium Wilt Pathogen of Banana, in Ecuador. Plants, 9(9), 1133. https://doi.org/10.3390/plants9091133
Martín, M. C., Leyva, L., Suárez, M. A., Pichardo, T., Caraballoso, I. B., & Capó, Y. A. (2021). Antifungal activity of Bacillus amyloliquefaciens against Fusarium oxysporum f. sp. cubense race 1. Agronomía Mesoamericana, 466-478. https://doi.org/10.15517/am.v32i2.39720
Maurhofer, M., Baehler, E., Notz, R., Martinez, V., & Keel, C. (2004). Cross talk between 2, 4-diacetylphloroglucinol-producing biocontrol pseudomonads on wheat roots. Applied and Environmental Microbiology, 70(4), 1990-1998. https://doi.org/10.1128/AEM.70.4.1990-1998.2004
Meldrum, R. A., Daly, A. M., Tran-Nguyen, L. T. T., & Aitken, E. A. B. (2013). The effect of surface sterilants on spore germination of Fusarium oxysporum f. sp. cubense tropical race 4. Crop protection, 54, 194-198. https://doi.org/10.1016/J.CROPRO.2013.08.014
Meyer, J. A., & Abdallah, M. A. (1978a). The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. Microbiology, 107(2), 319-328. https://doi.org/10.1099/00221287-107-2-319.
Meyer, J. M., Azelvandre, P., & Georges, C. (1992b). Iron metabolism in Pseudomonas: salicylic acid, a siderophore of Pseudomonas fluorescens CHA0. BioFactors (Oxford, England), 4(1), 23-27.
Mishra, A. K., & Baek, K. H. (2021). Salicylic acid biosynthesis and metabolism: a divergent pathway for plants and bacteria. Biomolecules, 11(5), 705. https://doi.org/10.3390/biom11050705
Mostert, D., Molina, A. B., Daniells, J., Fourie, G., Hermanto, C., Chao, C. P., ... & Viljoen, A. (2017). The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f. sp. cubense, in Asia. PLoS One, 12(7), e0181630. https://doi.org/10.1371/journal.pone.0181630
Naik, P. R., Sahoo, N., Goswami, D., Ayyadurai, N., & Sakthivel, N. (2008). Genetic and functional diversity among fluorescent pseudomonads isolated from the rhizosphere of banana. Microbial ecology, 56, 492-504. https://doi.org/10.1007/s00248-008-9368-9
Niwas, R., Chand, G., & Gupta, R. N. (2022). Fusarium wilt: a destructive disease of banana and their sustainable management. In Fusarium-An Overview of the Genus. IntechOpen. https://doi.org/10.5772/intechopen.101496
Odori, C., Ngaira, J., Kinyua, J., & Nyaboga, E. N. (2020). Morphological, genetic diversity and symbiotic functioning of rhizobia isolates nodulating cowpea (Vigna unguiculata L. Walp) in soils of Western Kenya and their tolerance to abiotic stress. Cogent Food & Agriculture, 6(1), 1853009. https://doi.org/10.1080/23311932.2020.1853009
Oktavioni, M., Winata, S. R., Syafriani, E., Syukriani, L., & Jamsari, J. (2020, April). Isolation of Chitinase B [ChiB] gene from Serratia plymutica strain UBCF_13. IOP Conference Series: Earth and Environmental Science, 497(1), 012020. https://doi.org/10.1088/1755-1315/497/1/012020
Parvin, W., Rahman, M. M., Govender, N. T., & Wong, M. Y. (2020). Identification, determination and quantification of indole-3-acetic acid produced by Pseudomonas aeruginosa UPMP3 and its effect on the growth of oil palm (Elaeis guineensis Jacq). World Journal of Agricultural Research, 8(3), 75-83. https://doi.org/10.12691/wjar-8-3-2
Patten, C. L., & Glick, B. R. (1996a). Bacterial biosynthesis of indole-3-acetic acid. Canadian journal of microbiology, 42(3), 207-220. https://doi.org/10.1139/m96-032
Patten, C. L., & Glick, B. R. (2002b). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Applied and environmental microbiology, 68(8), 3795-3801. https://doi.org/10.1128/AEM.68.8.3795-3801.2002
Patten, C. L., Blakney, A. J., & Coulson, T. J. (2013c). Activity, distribution and function of indole-3-acetic acid biosynthetic pathways in bacteria. Critical reviews in microbiology, 39(4), 395-415. https://doi.org/10.3109/1040841X.2012.716819
Peng, H., Zhang, P., Bilal, M., Wang, W., Hu, H., & Zhang, X. (2018). Enhanced biosynthesis of phenazine-1-carboxamide by engineered Pseudomonas chlororaphis HT66. Microbial cell factories, 17, 1-12.
Peñafiel-Jaramillo, M., Barrera-Álvarez, A. E., Torres-Navarrete, E. D., Canchignia-Martínez, H. F., Prieto-Encalada, H., & Morante-Carriel, J. (2016). Producción de ácido indol-3-acético por Pseudomonas veronii R4 y formación de raíces en hojas de vid “Thompson seedless” in vitro. Ciencia y Tecnología, 9, 31-36. https://doi.org/10.18779/cyt.v9i1.158
Prisa, D., Fresco, R., & Spagnuolo, D. (2023). Microbial biofertilisers in plant production and resistance: A review. Agriculture, 13(9), 1666. https://doi.org/10.3390/AGRICULTURE13091666
Qin, Y., Xie, X. Q., Khan, Q., Wei, J. L., Sun, A. N., Su, Y. M., ... & Xing, Y. X. (2022). Endophytic nitrogen-fixing bacteria DX120E inoculation altered the carbon and nitrogen metabolism in sugarcane. Frontiers in Microbiology, 13, 1000033. https://doi.org/10.3389/fmicb.2022.1000033
Rai, R., Srinivasamurthy, R., Dash, P. K., & Gupta, P. (2017). Isolation, characterization and evaluation of the biocontrol potential of Pseudomonas protegens RS-9 against Ralstonia solanacearum in Tomato. Indian Jorunal of Experimental Biology, 55, 595-603.
Ramaiah, N., Hill, R. T., Chun, J., Ravel, J., Matte, M. H., Straube, W. L., & Colwell, R. R. (2000). Use of a chiA probe for detection of chitinase genes in bacteria from the Chesapeake Bay. FEMS Microbiology Ecology, 34(1), 63-71. https://doi.org/10.1111/j.1574-6941.2000.tb00755.x
Ramette, A., Moënne-Loccoz, Y., & Défago, G. (2001). Polymorphism of the polyketide synthase gene phlD in biocontrol fluorescent pseudomonads producing 2, 4-diacetylphloroglucinol and comparison of PhlD with plant polyketide synthases. Molecular plant-microbe interactions, 14(5), 639-652. https://doi.org/10.1094/MPMI.2001.14.5.639
Ranjbar, R., Tabatabaee, A., Behzadi, P., & Kheiri, R. (2017). Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) genotyping of Escherichia coli strains isolated from different animal stool specimens. Iranian journal of pathology, 12(1), 25. https://doi.org/10.30699/ijp.2017.21506
Rijavec, T., & Lapanje, A. (2016). Hydrogen cyanide in the rhizosphere: not suppressing plant pathogens, but rather regulating availability of phosphate. Frontiers in microbiology, 7, 216209. https://doi.org/10.3389/fmicb.2016.01785
Rouhrazi, K., & Khodakaramian, G. (2015). Phenotypic and genotypic diversity of root-nodulating bacteria isolated from chickpea (Cicer arietinum L.) in Iran. Annals of microbiology, 65, 2219-2227. https://doi.org/10.1007/s13213-015-1062-9
Sarker, A., & Al-Rashid, J. (2013). Analytical protocol for determination of Indole 3 acetic acid (IAA) production by Plant Growth Promoting Bacteria (PGPB). Technical report of Quantification of IAA by microbes September: 3–5.
Satilmis, S., Toprak, N. U., Ilgın, C., & Soyletir, G. (2019). Evaluation of direct 16S rRNA PCR from clinical samples for bacterial detection in normally sterile body sites. The Journal of Infection in Developing Countries, 13(11), 978-983. https://doi.org/10.3855/jidc.11732
Shrivastava, U. P. (2017). Molecular diversity assessment of plant growth promoting rhizobacteria using denaturing gradient gel electrophoresis (DGGE) of 16s rRNA gene. International Journal of Applied Sciences and Biotechnology, 5(1), 72-80. https://doi.org/10.3126/ijasbt.v5i1.17029
Siamak, S. B., & Zheng, S. (2018). Banana Fusarium wilt (Fusarium oxysporum f. sp. cubense) control and resistance, in the context of developing wilt-resistant bananas within sustainable production systems. Horticultural Plant Journal, 4(5), 208-218. https://doi.org/10.1016/J.HPJ.2018.08.001
Singh, B. P. (2014). Genetic fingerprinting of antimicrobial fluorescent Pseudomonads associated with banana rhizosphere. Austin Journal of Biotechnology & Bioengineering, 1(2), 1-6.
Statgraphics Technologies (2019). Statgraphics Centurion Version 18 User ́s Manual. The Plains, VA, USA: Statgraphics Inc.
Suresh, P., Varathraju, G., Shanmugaiah, V., Almaary, K. S., Elbadawi, Y. B., & Mubarak, A. (2021). Partial purification and characterization of 2, 4-diacetylphloroglucinol producing Pseudomonas fluorescens VSMKU3054 against bacterial wilt disease of tomato. Saudi Journal of Biological Sciences, 28(4), 2155-2167. https://doi.org/10.1016/j.sjbs.2021.02.073
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, 30(12), 2725-29. https://doi.org/10.1093/molbev/mst197
Thangavelu, R., Gopi, M., Pushpakanth, P., Loganathan, M., Edwin Raj, E., Marimuthu, N., ... & Uma, S. (2021). First Report of Fusarium oxysporum f. sp. cubense VCG 0125 and VCG 01220 of Race 1 Infecting Cavendish Bananas (Musa sp. AAA) in India. Plant Disease, 105(4), 1215-1215. https://doi.org/10.1094/PDIS-09-20-2052-PDN
Vega-Celedón, P., Canchignia Martínez, H., González, M., & Seeger, M. (2016). Biosíntesis de ácido indol-3-acético y promoción del crecimiento de plantas por bacterias. Cultivos tropicales, 37, 33-39. https://doi.org/10.13140/RG.2.1.5158.3609
Verma, P., Chandra, P., Rai, A. K., Basak, N., Sundha, P., Sehrawat, A., ... & Yadav, R. K. (2023). Isolation, Screening and Evaluation of Biocontrol Potential of Rhizobacteria isolated from different agro-ecologies. Journal of Soil Salinity and Water Quality, 15(2), 229-241.
Versalovic, J. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods in Molecular and Cellular Biology, 5(1), 25-40.
Visca, P., Ciervo, A., Sanfilippo, V., & Orsi, N. (1993). Iron-regulated salicylate synthesis by Pseudomonas spp. Microbiology, 139(9), 1995-2001. https://doi.org/10.1099/00221287-139-9-1995
Were, E., Viljoen, A., & Rasche, F. (2023). Iron necessity for chlamydospore germination in Fusarium oxysporum f. sp. cubense TR4. BioMetals, 36(6), 1295-1306. https://doi.org/10.1007/s10534-023-00519-4.
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Scientia Agropecuaria

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The authors who publish in this journal accept the following conditions:
a. The authors retain the copyright and assign to the magazine the right of the first publication, with the work registered with the Creative Commons attribution license, which allows third parties to use the published information whenever they mention the authorship of the work and the First publication in this journal.
b. Authors may make other independent and additional contractual arrangements for non-exclusive distribution of the version of the article published in this journal (eg, include it in an institutional repository or publish it in a book) as long as it clearly indicates that the work Was first published in this journal.
c. Authors are encouraged to publish their work on the Internet (for example, on institutional or personal pages) before and during the review and publication process, as it can lead to productive exchanges and a greater and faster dissemination of work Published (see The Effect of Open Access).




