A computational analysis revealed BES1 transcription factor and β-amylase as crosstalk elements in Upland cotton species (Gossypium sp.)
DOI:
https://doi.org/10.17268/sci.agropecu.2024.033Keywords:
BAM, bioinformatics, BZR1, crosstalk, gene expression, transcription factorAbstract
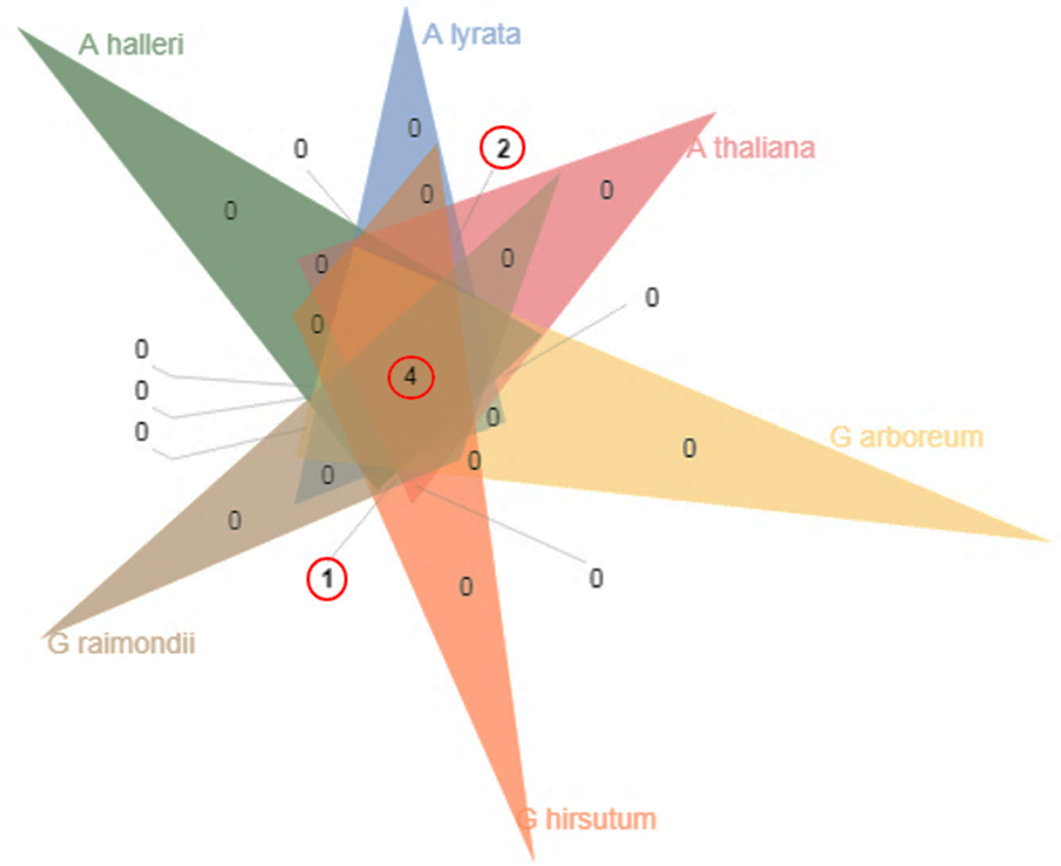
Cotton is a resilient and multipurpose crop, meeting major of the world’s textile needs while also yielding byproducts like edible oil and animal feed. Starch plays a crucial role in cotton fabric production. It enhances fabric strength by forming a protective film around cotton fibers, making them more resistant to wear and tear. BES1 (brassinosteroid insensitive 1) is a key regulator in brassinosteroid signaling. It controls thousands of target genes involved in development processes. Interestingly, two β-amylase proteins (BAM7 and BAM8) are part of the BES1 family, despite their primary function as β-amylases. β-Amylase (BAM) and BES1 are two gene families with functional and regulatory roles in controlling shoot growth and development by mediating brassinosteroid effects. They share similar domains and participate in various biological processes, tolerance and responses to stresses like salt and drought. In a computational analysis comparing Arabidopsis and Gossypium species, BAM and BES1 were characterized. BES1 genes were grouped into four clusters based on the comparison of the two species. Two clusters corresponded to BAM7 and BAM8, while the other two clusters were associated with BES1. The conserved nucleotide domain sequence is GCTGGATGG. Short tandem repeats include TG and TTG, which can serve as molecular markers. BES1 is specifically linked to cellulose and fiber production and holds promise as a candidate for plant selection and breeding in Gossypium (cotton).
References
Ahammed, G. J., Li, X., Liu, A., & Chen, S. (2020). Brassinosteroids in plant tolerance to abiotic stress. Journal of Plant Growth Regulation, 39 (4), 1451–1464. https://doi.org/10.1007/S00344-020-10098-0
Bailey, T. L., & Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings. International Conference on Intelligent Systems for Molecular Biology, 2, 28–36.
Campos, M. L., de Almeida, M., Rossi, M. L., Martinelli, A. P., Litholdo Junior, C. G., et al. (2009). Brassinosteroids interact negatively with jasmonates in the formation of anti-herbivory traits in tomato. Journal of Experimental Botany, 60(15), 4347–4361. https://doi.org/10.1093/jxb/erp270
Cao, X., Wei, Y., Shen, B., Liu, L., & Mao, J. (2024). Interaction of the transcription factors BES1/BZR1 in plant growth and stress response. International Journal of Molecular Sciences, 25(13), 6836. https://doi.org/10.3390/IJMS25136836
Gruszka, D., Srivastava, A. K., Yin, Y., & Kono, A. (2020). Updates on BES1/BZR1 regulatory networks coordinating plant growth and stress responses. Front. Plant Sci, 11, 617162. https://doi.org/10.3389/fpls.2020.617162
Huerta-Cepas, J., Serra, F., & Bork, P. (2016). ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Molecular Biology and Evolution, 33(6), 1635–1638. https://doi.org/10.1093/molbev/msw046
Jazayeri, S. M., Pooralinaghi, M., Villamar-Torres, R. O., & Cruzatty, L. C. G. (2018). An in silico comparative genomic report on transcription factors in three Arabidopsis species. Ciencia y Tecnología, 11(1), 1–9. https://doi.org/10.18779/cyt.v11i1.185
Jazayeri, S. M., Villamar-Torres, R. O., Zambrano-Vega, C., Cruzatty, L. C. G., Oviedo-Bayas, B., et al. (2020). Transcription factors and molecular markers revealed asymmetric contributions between allotetraploid Upland cotton and its two diploid ancestors. Bragantia, 79(1), 30-46. https://doi.org/10.1590/1678-4499.20190161
Jazayeri, S.M., García Cruzatty, L. C., Villamar-Torres, R., Cruzatty, L. C. G., & Villamar-Torres, R. (2019). Genomic comparison among three arabidopsis species revealed heavy metal responsive genes. Journal of Animal and Plant Sciences, 29(2).
John, M. E., & Crow, L. J. (1992). Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proceedings of the National Academy of Sciences of the United States of America, 89(13), 5769–73.
Kaplan, F., & Guy, C. L. (2004). beta-Amylase induction and the protective role of maltose during temperature shock. Plant Physiology, 135(3), 1674–84. https://doi.org/10.1104/pp.104.040808
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. https://doi.org/10.1093/molbev/msw054
Letunic, I., & Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research, 44(W1), W242–W245. https://doi.org/10.1093/nar/gkw290
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nature Biotechnology, 33(5), 524–530. https://doi.org/10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nature Genetics, 46(6), 567–572. https://doi.org/10.1038/ng.2987
MacGregor, E. A., Janecek, S., & Svensson, B. (2001). Relationship of sequence and structure to specificity in the alpha-amylase family of enzymes. Biochimica et Biophysica Acta, 1546(1), 1–20.
Manghwar, H., Hussain, A., Ali, Q., & Liu, F. (2022). Brassinosteroids (BRs) role in plant development and coping with different stresses. International Journal of Molecular Sciences, 23(3), 1012. https://doi.org/10.3390/IJMS23031012
Nolan, T., Chen, J., & Yin, Y. (2017). Cross-talk of Brassinosteroid signaling in controlling growth and stress responses. The Biochemical Journal, 474(16), 2641–2661. https://doi.org/10.1042/BCJ20160633
Rathore, K. S., Campbell, L. M., Sherwood, S., & Nunes, E. (2015). Cotton (Gossypium hirsutum L.). Methods in molecular biology (Clifton, N.J.), 1224, 11–23. https://doi.org/10.1007/978-1-4939-1658-0_2
Reinhold, H., Soyk, S., Simková, K., Hostettler, C., Marafino, J., et al. (2011). β-amylase-like proteins function as transcription factors in Arabidopsis, controlling shoot growth and development. The Plant Cell, 23(4), 1391–403. https://doi.org/10.1105/tpc.110.081950
Ryu, H., Cho, H., Bae, W., & Hwang, I. (2014). Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nature Communications, 5, 4138. https://doi.org/10.1038/ncomms5138
Shi, H., Li, X., Lv, M., & Li, J. (2022). BES1/BZR1 family transcription factors regulate plant development via brassinosteroid-dependent and independent pathways. International Journal of Molecular Sciences, 23(17), 10149. https://doi.org/10.3390/IJMS231710149/S1
Song, X., Ma, X., Li, C., Hu, J., Yang, Q., et al. (2018). Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genomics, 19(1), 1–15. https://doi.org/10.1186/S12864-018-4744-4
Todaka, D., Matsushima, H., & Morohashi, Y. (2000). Water stress enhances beta-amylase activity in cucumber cotyledons. Journal of Experimental Botany, 51(345), 739–745.
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nature Genetics, 44(10), 1098–1103. https://doi.org/10.1038/ng.2371
Wang, Y., Coleman-Derr, D., Chen, G., & Gu, Y. Q. (2015). OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Research, 43(W1), W78-W84. https://doi.org/10.1093/nar/gkv487
Wu, P., Song, X., Wang, Z., Duan, W., Hu, R., et al. (2016). Genome-wide analysis of the BES1 transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Growth Regulation, 80(3), 291–301. https://doi.org/10.1007/s10725-016-0166-y
Xie, L., Yang, C., & Wang, X. (2011). Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. Journal of Experimental Botany, 62(13), 4495–4506. https://doi.org/10.1093/jxb/err164
Yang, T., Li, H., Li, L., Wei, W., Huang, Y., Xiong, F., & Wei, M. (2023). Genome-wide characterization and expression analysis of α-amylase and β-amylase genes underlying drought tolerance in cassava. BMC Genomics, 24(1), 1–14. https://doi.org/10.1186/S12864-023-09282-9
Yasir, T. A., & Wasaya, A. (2021). Brassinosteroids Signaling pathways in plant defense and adaptation to stress. Plant Growth Regulators: Signalling under Stress Conditions, 197–206. https://doi.org/10.1007/978-3-030-61153-8_9
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., & Chory, J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell, 120(2), 249–259. https://doi.org/10.1016/j.cell.2004.11.044
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Scientia Agropecuaria

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The authors who publish in this journal accept the following conditions:
a. The authors retain the copyright and assign to the magazine the right of the first publication, with the work registered with the Creative Commons attribution license, which allows third parties to use the published information whenever they mention the authorship of the work and the First publication in this journal.
b. Authors may make other independent and additional contractual arrangements for non-exclusive distribution of the version of the article published in this journal (eg, include it in an institutional repository or publish it in a book) as long as it clearly indicates that the work Was first published in this journal.
c. Authors are encouraged to publish their work on the Internet (for example, on institutional or personal pages) before and during the review and publication process, as it can lead to productive exchanges and a greater and faster dissemination of work Published (see The Effect of Open Access).




