Foliar phytopathogenic fungi associated with the cultivation of Prunus persica L.: Morphological and molecular identification, and biological control with Trichoderma asperelloides
DOI:
https://doi.org/10.17268/sci.agropecu.2025.013Keywords:
Molecular analysis, biocontrol, plant confrontation, fungus, identification, T. asperelloidesAbstract
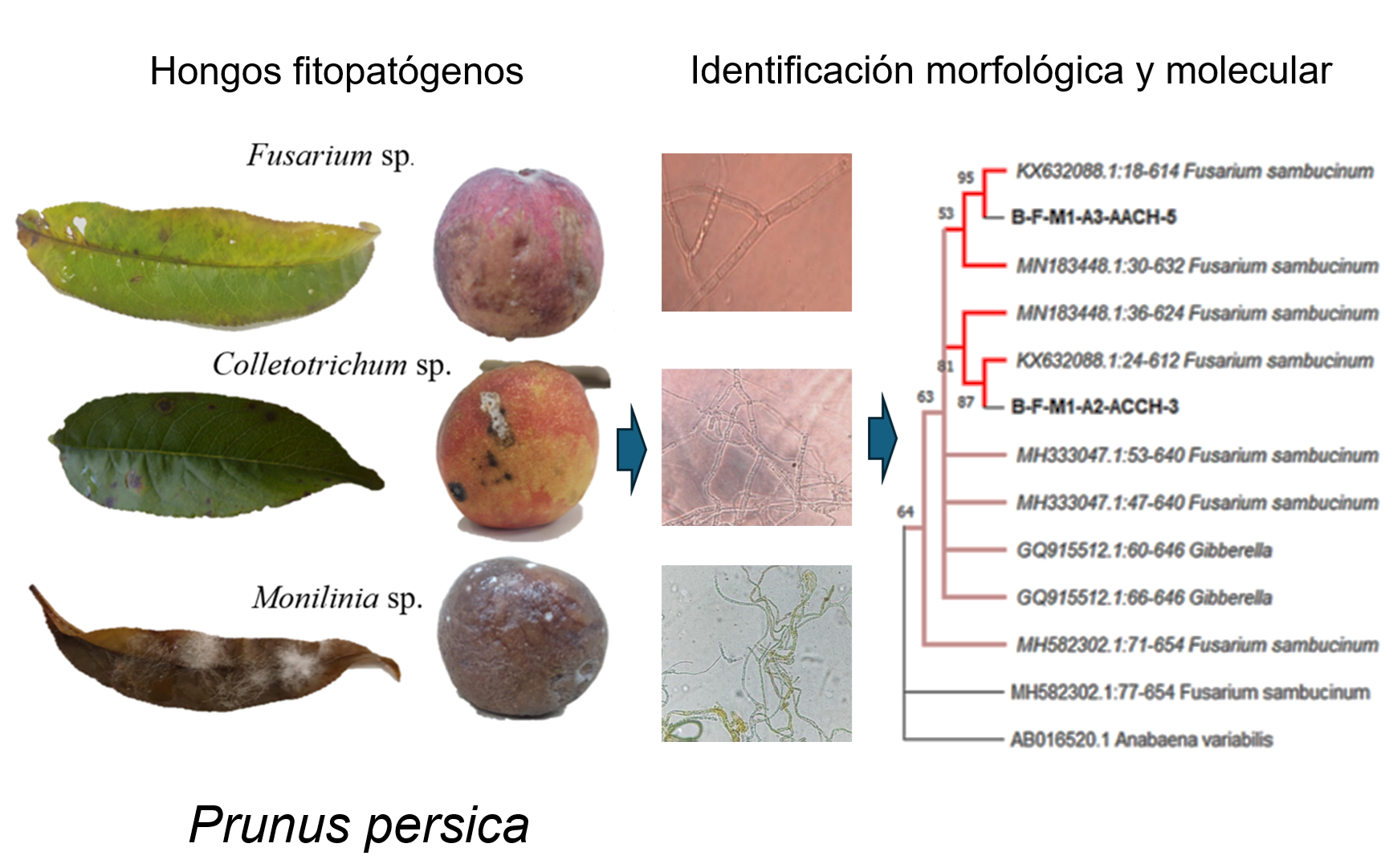
The presence of foliar phytopathogenic fungi causes severe damage to leave and fruits of peach (Prunus persica L.), in producing areas of southern Chihuahua, Mexico, which has caused a decrease in production by 30%. The objective of this work was to identify morphologically and molecularly the foliar phytopathogenic fungi associated with the peach tree crop, evaluating the pathogenicity in one-year-old plants against Trichoderma asperelloides. Leaves with brown and brown lesions were collected from mummified fruits on the plant from 19 commercial peach orchards distributed in three municipalities of regional and national production in the State of Chihuahua, Mexico. Fungal identification of four representative isolates was performed using morphological methods. characterization and phylogenetic analysis based on the internal transcribed spacer region (ITS1 and ITS4) of ribosomal DNA, part of the translation elongation factor 1-alpha (TEF) a second secondary primer for each of the genera for, Collectotrichum ACT-512F and ACT-583R, Fusarium with EF1. For plant confrontations, a concentration of 1x106 conidia was inoculated. mL-1 of pathogens such as T. asperelloides, evaluating leaf diameter and length, height, severity and incidence. It was possible to identify the presence of Fusarium sambucinum, Collectotrichum gleosporoides and Monilinia frutícola, in addition the B-F-M1-A2-ACCH-3 strain of F. Sambucinum obtained the highest values in the inhibition of the response variables and in severity Monilinia frutícola presented 61.23% as the highest value. It is recommended to use strain 3 as a biological control of foliar phytopathogens.
References
Agu, K. C., & Chidozie, C. P. (2021). An improved slide culture technique for the microscopic identification of fungal species. International Journal of Trend in Scientific Research and Development, 6(1), 243-254.
Almaraz-Sánchez, A., Ayala-Escobar, V., Tlatilpa-Santamaría, I. F., & Nieto-Angel, D. (2018). Fusarium sambucinum Fuckel agente causal de la pudrición de frutos de chile manzano (Capsicum pubescens) en México. Revista Mexicana de Fitopatología, 37(1). https://doi.org/10.18781/r.mex.fit.1810-2
Arafat, K. H., Hassan, M., & Hussein, E. A. (2021). Detection, disease severity and chlorophyll prediction of date palm leaf spot fungal diseases. New Valley Journal of Agricultural Science, 1(2), 98-110.
https://doi.org/10.21608/nvjas.2022.110022.1027
Baltazar, E., Rodrigues, S., Ares, A., Camelo, A., Brandão, I., et al. (2023). Morphological, Molecular and Genomic Identification and Characterisation of Monilinia fructicola in Prunus persica from Portugal. Agronomy, 13(6), 1493. https://doi.org/10.3390/agronomy13061493
Battistini, G., Gazzetti, K., & Collina, M. (2022). A New Approach: Determining cyt b G143A Allele Frequency in Zymoseptoria tritici by Digital Droplet PCR. Biology, 11(2), 240. https://doi.org/10.3390/biology11020240
Boukaew, S., Chumkaew, K., Petlamul, W., Srinuanpan, S., Nooprom, K., & Zhang, Z. (2024). Biocontrol effectiveness of Trichoderma asperelloides SKRU-01 and Trichoderma asperellum NST-009 on postharvest anthracnose in chili pepper. Food Control, 163, 110490. https://doi.org/10.1016/j.foodcont.2024.110490
Da Silva Neto, J. A., De Queiroz Ambrósio, M. M., Araújo, M. B. M., Da Silva, R. M., Pinto, P. S. L., & Holanda, I. S. A. (2022). Morphological, molecular and pathogenic characterization of Colletotrichum gloeosporioides isolated from mango. Revista Caatinga, 35(3), 514-527. https://doi.org/10.1590/1983-21252022v35n302rc
Dini, M., Raseira, M., Scariotto, S., Marchi, P., & Mello-Farias, P. (2021). Research article peach phenological characters: heritability, maternal effect and correlation with brown rot. Genetics and Molecular Research, 20(1). https://doi.org/10.4238/gmr18684
Dong, J., Shi, H., Wu, Y., Yang, L., Zhu, F., & Ji, Z. (2023). Identification and pathogenicity analysis of Fusarium spp. on peach in China. BMC Microbiology, 23, 211. https://doi.org/10.1186/s12866-023-02958-y
Edgar, R. C. (2004). Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792-1797. https://doi.org/10.1093/nar/gkh340
Gerardo-Lugo, S. S., Tovar-Pedraza, J. M., Maharachchikumbura, S. S., Apodaca-Sánchez, M. A., Correia, K. C., et al. (2020). Characterization of Neopestalotiopsis species associated with mango grey leaf spot disease in Sinaloa, Mexico. Pathogens, 9(10), 788. https://doi.org/10.3390/pathogens9100788
Gununu, P. R., Munhuweyi, K., Obianom, P. C., & Sivakumar, D. (2019). Assessment of eleven South African peach cultivars for susceptibility to brown rot and blue mould. Scientia Horticulturae, 254, 1-6. https://doi.org/10.1016/j.scienta.2019.04.067
Grano-Maldonado, M. I., Ramos-Payan, R., Rivera-Chaparro, F., Aguilar-Medina, M., Romero-Quintana, J. G., Rodríguez-Santiago, A., & Nieves-Soto, M. (2021). First Molecular Characterization of Colletotrichum sp. and Fusarium sp. Isolated from Mangrove in Mexico and the Antagonist Effect of Trichoderma harzianum as an Effective Biocontrol Agent. Plant Pathology Journal, 37(5), 465-475. https://doi.org/10.5423/ppj.oa.03.2021.0048
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series, 41(49), 95-98.
Heflish, A. A., Abdelkhalek, A., Al-Askar, A. A., & Behiry, S. I. (2021). Protective and Curative Effects of Trichoderma asperelloides Ta41 on Tomato Root Rot Caused by Rhizoctonia solani Rs33. Agronomy, 11(6), 1162. https://doi.org/10.3390/agronomy11061162
Huang, R., Sun, W., Wang, L., Li, Q., Huang, S., et al. (2021). Identification and characterization of Colletotrichum species associated with anthracnose disease of banana. Plant Pathology, 70(8), 1827-1837. https://doi.org/10.1111/ppa.13426
Huilotl-Luna, M. L., Gil-Muñoz, A., Hernández-Romero, E., López, P. A., & Martínez-Carrera, D. C. (2024). Manejo de la pudrición café (Monilinia fructicola) del duraznero por productores de la Sierra Nevada de Puebla. Agricultura Sociedad y Desarrollo, 21(4). https://doi.org/10.22231/asyd.v21i4.1679
Iñiguez-Moreno, M., Sandoval-Contreras, T., Ragazzo-Sánchez, J. A., & Calderón-Santoyo, M. (2023). Modelado del crecimiento de hongos fitopatógenos aislados de frutos de aguacate Hass. Acta de Ciencia en Salud, 20(7), 1-5.
Iqbal, S., Abbas, A., Mubeen, I., Sathish, M., Razaq, Z., Mubeen, M., Kamran, M., Haroon, M., Syed, S., Naqvi, S., & Ahmed, M. A. A. (2022). Taxonomy, distribution, epidemiology, disease cycle and management of brown rot disease of peach (Monilinia spp.). Notulae Botanicae Horti Agrobotanici Cluj-Napoca, 50(1), 12630. https://doi.org/10.15835/nbha50112630
Karlsson, I., Edel-Hermann, V., Gautheron, N., Durling, M. B., Kolseth, A. K., et al. (2016). Genus-specific primers for study of Fusarium communities in field samples. Applied and environmental microbiology, 82(2), 491-501. https://doi.org/10.1128/AEM.02748-15
Lanfear, R., Calcott, B., Ho, S. y. W., & Guindon, S. (2012). PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Molecular Biology And Evolution, 29(6), 1695-1701. https://doi.org/10.1093/molbev/mss020
Luo, C., Schnabel, G., Hu, M., & De Cal, A. (2022). Global distribution and management of peach diseases. Phytopathology Research, 4(1). https://doi.org/10.1186/s42483-022-00134-0
Mannai, S., & Boughalleb-M’Hamdi, N. (2022). In vitro and in planta potential effect of some indigenous antagonists against Fusarium and pythiaceous species associated with peach seedlings decline. Egyptian Journal of Biological Pest Control, 32(1). https://doi.org/10.1186/s41938-022-00540-8
Manganaris, G. A., Minas, I., Cirilli, M., Torres, R., Bassi, D., & Costa, G. (2022). Peach for the future: A specialty crop revisited. Scientia Horticulturae, 305, 111390. https://doi.org/10.1016/j.scienta.2022.111390
Moreno, L. J. P., Prado, D. A. A., & Herrera, S. Y. A. (2018). Hongos fitopatógenos asociados a enfermedades foliares de Cattleya, Miltoniopsis y Oncidium en víveros de fusagasugá (Cundinamarca, Co). Revista de Fitopatología Colombiana, 42(1), 13-16.
Özer, N., Uzun, H. İ., Aktürk, B., Özer, C., Akkurt, M., & Aydın, S. (2021). Resistance assessment of grapevine leaves to downy mildew with sporulation area scoring. European Journal Of Plant Pathology, 160(2), 337-348. https://doi.org/10.1007/s10658-021-02247-2
Ramírez-Cariño, H. F., Guadarrama-Mendoza, P. C., Sánchez-López, V., Cuervo-Parra, J. A., Ramírez-Reyes, T., Dunlap, C. A., & Valadez-Blanco, R. (2020). Biocontrol of Alternaria alternata and Fusarium oxysporum by Trichoderma asperelloides and Bacillus paralicheniformis in tomato plants. Antonie Van Leeuwenhoek, 113(9), 1247-1261. https://doi.org/10.1007/s10482-020-01433-2
Ren, F., Dong, W., & Yan, D. (2019). Organs, cultivars, soil, and fruit properties affect structure of endophytic mycobiota of pinggu peach trees. Microorganisms, 7(9), 322. https://doi.org/10.3390/microorganisms7090322
Rios-Hernández, T. A., Uc-Varguez, A., & Evangelista-Martínez, Z. (2021). Biological control of Fusarium oxysporum causal agent of gladiolus corm rot by streptomycetes. Revista Mexicana de Fitopatología, 39(3). https://doi.org/10.18781/r.mex.fit.2105-3
Silva-Neto, J. A. D., Ambrósio, M. M. D. Q., Araújo, M. B. M., Silva, R., Pinto, P. S. L., & Holanda, I. S. A. (2022). Morphological, molecular and pathogenic characterization of Colletotrichum gloeosporioides isolated from mango. Revista Caatinga, 35, 514-527. https://doi.org/10.1590/1983-21252022v35n302rc
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22(21), 2688-2690. https://doi.org/10.1093/bioinformatics/btl446
Statistical Analysis Software. (SAS). (2012). Users’ Guide Statistics Version 9.4. SAS Institute Inc., Cary. https://support.sas.com/software/94/
Tan, Q., Schnabel, G., Chaisiri, C., Yin, L. F., Yin, W. X., & Luo, C. X. (2022). Colletotrichum species associated with peaches in China. Journal of Fungi, 8(3), 313. https://doi.org/10.3390/jof8030313
Udhayakumar, R., Usharani, S., & Muthukumar, A. (2019). Pathogenicity variation, morphological and cultural characteristic of Colletotrichum gloeosporioides isolates. Plant Archives, 19(1), 425-430.
Voglmayr, H, Akulov, O. Y., & Jaklitsch, W. M. (2016). Reassessment of Allantonectria, phylogenetic position of Thyronectroidea, and Thyronectria caraganae sp. nov. Mycological Progress, 15(9), 921–937. https://doi.org/10.1007/s11557-016-1218-4
Wang, Q., Fan, K., Li, D., Han, C., Qu, Y., Qi, Y., & Wu, X. (2020). Identification, Virulence and Fungicide Sensitivity of Colletotrichum gloeosporioides s.s. Responsible for Walnut Anthracnose Disease in China. Plant Disease, 104(5), 1358-1368. https://doi.org/10.1094/pdis-12-19-2569-re
White, T., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. En Elsevier eBooks (pp. 315-322). https://doi.org/10.1016/b978-0-12-372180-8.50042-1
Yikilmazsoy, G., & Tosun, N. (2021). Characterization of Fusarium sambucinum isolates associated with potato dry rot and evaluation of cultivar susceptibility and fungicides. Turkish Journal of Agriculture and Forestry, 45(2), 222-233. https://doi.org/10.3906/tar-2006-100
Yin, L., Zhang, S., Du, J., Wang, X., Xu, W., & Luo, C. (2021). Monilinia fructicola on loquat: An old pathogen invading a new host. Journal Of Integrative Agriculture, 20(7), 2009-2014. https://doi.org/10.1016/s2095-3119(20)63375-5
Yin, L. F., Mo, W., Guo, D. Y., & Luo, C. X. (2020). First Report of Brown Rot of Prunus mume Caused by Monilinia fructicola in China. Plant Disease, 104(4), 1253. https://doi.org/10.1094/pdis-09-19-1847-pdn
Zhan, Q., Wang, N., Jin, S., Tan, R., Jiang, Q., & Wang, Y. (2018) ProbPFP: A multiple sequence alignment algorithm combining partition function and hidden markov model with particle swarm optimization. IEEE International conference on bioinformatics and biomedicine (BIBM) 1290-1295. https://doi.org/10.1109/BIBM.2018.8621220
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Scientia Agropecuaria

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The authors who publish in this journal accept the following conditions:
a. The authors retain the copyright and assign to the magazine the right of the first publication, with the work registered with the Creative Commons attribution license, which allows third parties to use the published information whenever they mention the authorship of the work and the First publication in this journal.
b. Authors may make other independent and additional contractual arrangements for non-exclusive distribution of the version of the article published in this journal (eg, include it in an institutional repository or publish it in a book) as long as it clearly indicates that the work Was first published in this journal.
c. Authors are encouraged to publish their work on the Internet (for example, on institutional or personal pages) before and during the review and publication process, as it can lead to productive exchanges and a greater and faster dissemination of work Published (see The Effect of Open Access).




