Management Information system for hemoparasitosis detection in canines through machine learning techniques
Sistema de información gerencial para la detección de hemoparasitosis en caninos mediante técnicas de aprendizaje automático
Wilson Molina-Oleas1; Maritza Aguirre-Munizaga1, 2 *; Anthony Tatez-Luzardo1; Erwin Marin-Fajardo1
1 Computación, Facultad de Ciencias Agrarias, Universidad Agraria del Ecuador, Av.25 de Julio y Pio Jaramillo, Guayaquil, Ecuador.
2 Instituto de Investigación, Universidad Agraria del Ecuador, Av.25 de Julio y Pio Jaramillo, Guayaquil, Ecuador.
ORCID de los autores:
W. Molina-Oleas: https://orcid.org/0000-0003-2344-5064 M. Aguirre-Munizaga: https://orcid.org/0000-0003-0708-1811
A. Tatez-Luzardo: https://orcid.org/0009-0003-4465-2115 E. Marín-Fajardo https://orcid.org/0009-0008-8014-187X
ABSTRACT
This study presents the development and implementation of a web-based and mobile Management Information System (MIS) for the early detection and diagnosis of hemoparasitosis in canines, using machine learning algorithms. The system was deployed at Animal Place, a veterinary clinic in Guayaquil, in response to the increasing number of clinical cases associated with hemoparasitic infections. The platform integrates multiple clinical management modules, including a dedicated diagnostic module powered by a decision tree classification algorithm (J48), which achieved an accuracy of 96.4% based on a training set of 746 symptomatological records. The mobile application complements the web system by allowing users to monitor their pets’ clinical data and interact with an integrated chatbot assistant developed with DialogFlow. For system development, the Extreme Programming (XP) methodology was employed to ensure iterative progress, client feedback, and continuous testing. In parallel, the Knowledge Discovery in Databases (KDD) methodology was used to structure and preprocess clinical data, enabling the identification of key variables relevant to hemoparasitosis and training of the predictive model. Usability testing was conducted with 10 participants, including veterinary staff and administrative personnel. Results showed high levels of satisfaction (mean score = 4.5 on a 5-point scale), particularly regarding system functionality, ease of use, and interface clarity. These findings highlight the system's relevance, practicality, and user acceptance in a clinical setting. In conclusion, the proposed MIS successfully automates critical veterinary processes such as client registration, clinical history management, and disease detection, while significantly enhancing the accuracy and efficiency of early hemoparasitosis diagnosis in dogs. This research points out the transformative effects of machine learning and intelligent digital tools in veterinary practice and provides a scalable model for similar applications in animal health.
Keywords: Veterinary diagnostics; machine learning; clinical decision-making; information management systems.
RESUMEN
Este estudio presenta el desarrollo e implementación de un Sistema de Información Gerencial (SIG) web y móvil para la detección temprana y el diagnóstico de hemoparasitosis canina, mediante algoritmos de aprendizaje automático. El sistema se implementó en Animal Place, una clínica veterinaria en Guayaquil, en respuesta al creciente número de casos clínicos asociados con infecciones hemoparasitarias. La plataforma integra múltiples módulos de gestión clínica, incluyendo un módulo de diagnóstico específico impulsado por un algoritmo de clasificación de árboles de decisión (J48), que alcanzó una precisión del 96,4% con base en un conjunto de entrenamiento de 746 registros sintomatológicos. La aplicación móvil complementa el sistema web, permitiendo a los usuarios monitorear los datos clínicos de sus mascotas e interactuar con un asistente de chatbot integrado desarrollado con DialogFlow. Para el desarrollo del sistema, se empleó la metodología de Programación Extrema (XP) para asegurar el progreso iterativo, la retroalimentación del cliente y la realización de pruebas continuas. Paralelamente, se empleó la metodología de Descubrimiento de Conocimiento en Bases de Datos (KDD) para estructurar y preprocesar los datos clínicos, lo que permitió la identificación de variables clave relevantes para la hemoparasitosis y el entrenamiento del modelo predictivo. Se realizaron pruebas de usabilidad con 10 participantes, incluyendo personal veterinario y administrativo. Los resultados mostraron altos niveles de satisfacción (puntuación media = 4,5 en una escala de 5 puntos), especialmente en cuanto a la funcionalidad del sistema, la facilidad de uso y la claridad de la interfaz. Estos hallazgos resaltan la relevancia, la practicidad y la aceptación del sistema en el ámbito clínico. En conclusión, el SIG propuesto automatiza con éxito procesos veterinarios críticos, como el registro de clientes, la gestión de historiales clínicos y la detección de enfermedades, a la vez que mejora significativamente la precisión y la eficiencia del diagnóstico precoz de hemoparasitosis canina. Esta investigación destaca los efectos transformadores del aprendizaje automático y las herramientas digitales inteligentes en la práctica veterinaria y proporciona un modelo escalable para aplicaciones similares en la salud animal.
Palabras clave: diagnóstico veterinario; aprendizaje automático; decisión clínica; sistemas de gestión de información.
1. Introduction
Currently, domestic animals are increasingly exposed to a wide range of pathogenic microorganisms, including protozoa, bacteria, viruses, and fungi, posing a significant threat to animal health. Among these threats, hemoparasitic infections represent a major concern in veterinary medicine due to their prevalence, clinical complexity, and potential zoonotic implications (Qureshi et al., 2023). Hemoparasites are microscopic protozoan organisms that inhabit and reproduce within the bloodstream, generating various pathogenic effects that compromise the health of affected animals(da Silveira et al., 2011). If left untreated, these infections can lead to severe clinical conditions and, in some cases, pose risks to human health, thereby constituting a broader public health issue.
In canines, the most common clinical manifestations of hemoparasitosis include anemia, jaundice, fever, hind limb weakness, and thrombocytopenia (Fiuza Lemos Ferreira, 2022). The timely detection and diagnosis of such conditions are critical to ensure effective treatment and prevent complications. However, the increasing incidence of hemoparasitic diseases, partially driven by environmental factors such as climate change, demands the incorporation of advanced technological solutions in veterinary practice.
This research was conducted at Animal Place, a veterinary clinic located in Guayaquil, Ecuador, which attends to approximately 25 patients per week and is staffed by two veterinarians and one administrative professional. The clinic offers medical consultation, grooming services, and a pharmacy. Given the high frequency of hemoparasitic cases observed, the main objective of this study was to implement a web and mobile-based system for the detection and diagnosis of canine hemoparasitosis through machine learning algorithms. This technological integration seeks to enhance diagnostic accuracy, optimize clinical workflows, and support evidence-based veterinary decision-making (Ybanez et al., 2018).
In Ecuador, both felines and canines have become integral companions within household environments, particularly in urban and suburban areas. This growing bond between humans and domestic animals has driven initiatives to promote responsible pet ownership through legislative frameworks. One such example is the Organic Animal Welfare Law (LOBA), established under the Ecuadorian Constitution, which aims to “guarantee coexistence in harmonious and functional societies, where the exercise of the rights of citizens, nature, and the animals that comprise it are respected” (Hernández Bustos & Fuentes Terán, 2018).
Despite these legal advances, in rural, peri-urban, and marginalized communities, dogs are often still perceived primarily as functional animals used for home protection. This limited perspective has contributed to an unregulated increase in the canine population, often resulting in abandonment and exposure to harsh environmental conditions circumstances that represent a growing public health concern (Ord & Lobo, 2015).
One of the most pressing health threats to free-roaming and domestic dogs alike is hemoparasitosis, a group of globally prevalent diseases caused by blood-borne parasites that infect and consume red blood cells. These infections can lead to severe clinical outcomes, and in the absence of early diagnosis and proper treatment, they may result in death. In Ecuador, the most frequently reported hemoparasitic diseases in dogs are babesiosis and ehrlichiosis, both of which are transmitted predominantly by ticks of the Rhipicephalus genus (Adams et al., 2016).
Babesiosis is caused by Babesia spp., leading to anemia, fever, lethargy, and appetite loss. Ehrlichiosis, caused by Ehrlichia canis, targets white blood cells and produces similar symptoms, including weight loss, limping, and coagulopathies (Chua et al., 2020). Both diseases are typically diagnosed through laboratory tests such as blood smear microscopy, indirect immunofluorescence (IFI), and polymerase chain reaction (PCR). However, the cost, availability, and time constraints of these methods limit their widespread application in low-resource settings.
Given this context, the development of technological tools that support early, accurate, and accessible diagnosis of hemoparasitic infections in dogs becomes not only a veterinary necessity but also a public health priority (Karasová et al., 2022). The interpretation of diagnostic results necessitates a comprehensive understanding of the limitations and specificities of each test, as well as consideration of the animal's geographical location and travel history. Conside-ring these challenges, this paper aims to provide a comprehensive overview of the detection and diagnosis of canine hemoparasitosis, with a focus on the most prevalent etiological agents, diagnostic methodologies, and interpretative considerations.
2. Methodology
2.1 Research design
This research adopts a qualitative approach aimed at deeply understanding and exploring the detection of hemoparasitosis in canines within a veterinary clinical setting. The study involved data collection through semi-structured interviews that fostered active engagement between the resear-chers and the veterinary staff, facilitating the extraction of experiential and contextual know-ledge relevant to current diagnostic practices.
The researchers employed the Extreme Programming (XP) methodology to guide the development of an efficient web and mobile-based system for detecting hemoparasitosis. This agile software development framework enabled the iterative and incremental construction of the system while ensuring continuous feedback and adaptation to user needs. In parallel, qualitative data collection techniques, particularly interviews with experienced veterinarians, were conducted to obtain detailed insights into diagnostic proce-dures, challenges in the field, and the specific informational and operational requirements of veterinary professionals.
Content analysis was applied to examine the qualitative data systematically, identifying recurring patterns, themes, and conceptual categories related to clinical manifestations, diagnostic practices, and risk factors associated with hemoparasitic infections in dogs. This analysis provided a comprehensive understanding of the problem space and informed the design decisions of the technological solution.
Data collection and analysis were conducted rigorously and systematically, ensuring the credibility and reliability of the findings. The interpretation of results led to the generation of new perspectives, the identification of oppor-tunities for improvement in current diagnostic workflows, and the formulation of practical recommendations for the design and functionality of the information system.
This study integrates two complementary research types: documentary research and applied re-search with a qualitative orientation. Documentary research facilitated the examination of historical clinical records, allowing the identification of symptom patterns in previously diagnosed cases. Applied research, on the other hand, focused on developing a practical and context-sensitive technological solution.
Grounded in the principles of translational research, it bridges theoretical knowledge with the development of real-world applications. Through qualitative methods and stakeholder engagement, the study yielded valuable information on symptomatic indicators, diagnostic limitations, and end-user expectations, ultimately guiding the creation of a system tailored to the actual needs of veterinary practice.
2.2 Data analysis procedures
The Knowledge Discovery in Databases (KDD) methodology served as the analytical foundation for this research, providing a structured process for identifying meaningful patterns and extracting relevant knowledge from clinical data. As noted (Górecka et al., 2025) "The KDD process, on which most artificial intelligence projects that involve learning from existing data are based", plays a critical role in data-driven decision-making. Within the framework of this methodology, key variables indicative of hemoparasitosis were identified from the symptomatological records, allowing the training of a detection and diagnosis algorithm tailored to veterinary applications (Serin & Körez, 2025).
The KDD process was implemented through the following five stages:
2.2.1 Selection
In this initial phase, the relevant objectives and scope of knowledge discovery were defined according to the needs identified in the veterinary setting (Serin & Körez, 2025). Symptom records from the clinic were selected as the primary data source, as they represented clinically validated cases relevant to the detection of hemo-parasitosis. These records formed the basis for the machine learning algorithm, which delivers diagnostic results based on veterinarian selected symptoms within the web system.
2.2.2 Preprocessing
In order to find and eliminate records with null, duplicate, or inconsistent values, data prepro-cessing entailed a cooperative review with a knowledgeable veterinarian. The objective was to keep only the symptoms that were most important for diagnosis and to remove any redundant or unnecessary information. The resulting dataset contained markers linked to the absence of infection, parasite-specific symptoms, and general indicators of hemoparasitosis. This cleaned-up dataset was ready for model training and bulk upload.
2.2.3 Data transformation
After the data was cleaned, the dataset was optimized for algorithmic processing using trans-formation techniques. To improve the interpreta-bility of features across cases, these included normalization and discretization. At this point, variables like the breed of the dog, the date of symptom registration, specific symptoms, and associated diagnostic results were all included in the final dataset. A dataset of 746 instances was obtained by compiling the data into a structured CSV file, which was then used to train the machine learning model (Kraleva et al., 2018).
2.2.4 Machine learning model training
In the fourth stage of the KDD process, data mining techniques were applied to uncover hidden patterns within the transformed dataset and to build a predictive model capable of estimating target variables specifically, identifying which symptoms are most strongly associated with the presence of hemoparasitic infections in dogs. This stage involved the implementation of machine learning algorithms using the Weka software tool, where the CSV dataset was processed to test various classification models and determine the most suitable algorithm for the task (Kraleva et al., 2018).
Several machine learning algorithms were evaluated, including Bayesian Networks, logistic regression, and various decision tree-based methods. The results demonstrated that decision trees provided the highest predictive accuracy, with the J48 algorithm achieving a precision rate of 96.4%, followed by RandomTree (95.4%) and RandomForest (94.2%). These models were assessed based on accuracy, number of instan-ces processed, and prediction time. The decision tree was generated using symptomatological data collected from clinical records, incorporating varia-bles such as canine symptoms and diagnostic outcomes. This model formed the basis of the inference engine later integrated into the web-based system (Aguirre-Munizaga et al., 2025a).
Table 1 shows a summary of the performance metrics from the decision tree algorithms that were tested in 746 instances.
Table 1
Accuracy and prediction time of decision tree algorithms
Algorithm | Accuracy | Prediction time (seconds) |
J48 | 96.4% | 0.01 s |
RandomTree | 95.4% | 0.01 s |
RandomForest | 94.2% | 0.02 s |
LMT | 85.3% | 0.02 s |
DecisionStump | 61.0% | 0.04 s |
HoeffdingTree | 88.0% | 0.02 s |
REPTree | 85.0% | 0.02 s |
The superior performance of the J48 decision tree model made it the preferred choice for integration into the veterinary diagnostic system, ensuring both reliability and speed in the identification of hemoparasitosis (Dai & Ji, 2014).
2.3 System development methodology
The development of the system followed an agile and modular approach, utilizing technologies aligned with current best practices in web and mobile application engineering (Elasan & Yilmaz, 2025). The system was designed to support clini-cal management and disease diagnosis workflows in veterinary settings, ensuring scalability, securi-ty, and integration with intelligent diagnostic tools.
2.3.1 Web application development
The web platform was developed using the .NET Core framework, with the C# programming language employed for coding its core modules. C# was selected due to its robustness, flexibility, and suitability for enterprise level web appli-cations, particularly when combined with the .NET ecosystem, which facilitates secure and high-performance deployments (Aguirre-Munizaga et al., 2025b). The backend system interacts with a PostgreSQL database, where all essential tables and relational models are defined. An API layer was developed to manage client-server communication, utilizing HTTP POST and GET methods through specific endpoints corres-ponding to various data operations. This architecture ensures modularity and facilitates future system expansion.

Figure 1. Main navigation menu of the Animal Place veterinary System.
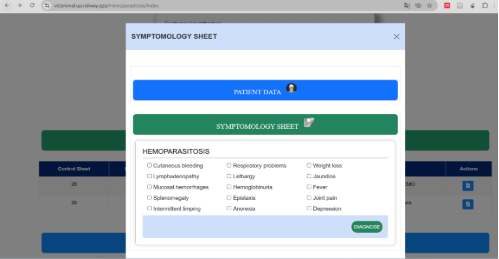
Figure 2. Symptomatology sheet interface for canine hemoparasitosis diagnosis.
For the diagnostic model, ML.NET was used to train and integrate the machine learning algorithm into the system. After preprocessing and selecting the appropriate classification algorithm, the model was trained using a dataset consisting of 746 instances (representing 60% of the total dataset). Performance testing was conducted to compare the system's prediction accuracy against the outcomes obtained during the earlier data mining phase in Weka.
2.3.2 Mobile application development
The mobile version of the system was developed using Android Studio, primarily utilizing Java for the application structure and Kotlin for functional modules. Within the Android manifest, user permissions were established to ensure secure access and proper data handling. The application includes core features such as user profile visualization, clinical history management, and a chatbot interface to support veterinary assistance.
The chatbot was implemented using DialogFlow, an open-source framework for developing conver-sational agents. DialogFlow operates through two main components: Natural Language Understan-ding (NLU), which enables the system to interpret user input, and Core, which is responsible for generating appropriate responses based on contextual data (Haryanto & Saefurrahman, 2024).
This architecture allows for dynamic, context-aware interactions with users.
Integration with the Android mobile application was achieved using Retrofit and Volley libraries, which support asynchronous HTTP POST requests and efficient communication with the chatbot server. We developed the conversational interface using RecyclerView, a component that facilitates the fluid visualization of dialogues, including user input and system responses. This interface is supported by a secure and modular API layer, allowing seamless exchange of clinical and diagnostic data between the mobile application and backend services.
Figure 3 presents a conceptual overview of the chatbot implementation, illustrating the interaction between the DialogFlow framework and the mobile interface. The structure ensures both technical robustness and user-centered design, facilitating intuitive and responsive user experiences in veterinary clinical environments.
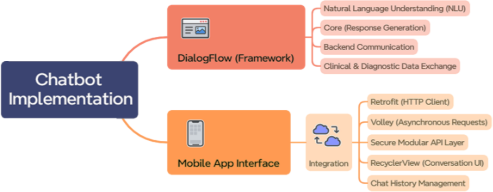
Figure 3. Chatbot implementation.
The API’s structure and endpoints are fully documented using Swagger, as illustrated in Figures 4 and 5. Figure 4 highlights endpoints for managing clients and hemoparasitosis diagnostics, while Figure 5 presents endpoints related to machine learning integration, QR code generation, breed classification, and diagnostic reporting. Together, these components ensure reliable interoperability between the mobile app and the backend system, supporting a robust and efficient veterinary diagnostic workflow.

Figure 4. API documentation for client and diagnostic modules.
3. System evaluation and usability testing
The study's results are shown through a mix of descriptive analysis, machine learning perfor-mance metrics, and validation of system functio-nality. To make things easier to understand and compare, quantitative results like algorithm accuracy rates and system test results are shown in tables. We also talk about qualitative observations from usability and interface testing to give you an idea of how users interact with the system and how it works in real clinical settings. Combining these points of view makes sure that both the technical performance and practical usefulness of the system that was made are fully evaluated (Niknejad et al., 2020).
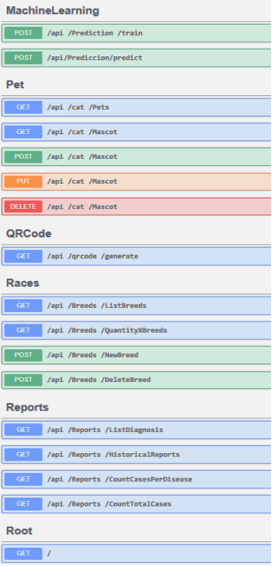
Figure 5. API endpoints for machine learning, QR code generation, breed classification, and reporting.
3.1 Unit testing
Unit testing was conducted during the system development process to verify the correct functionality of individual components before their integration into the broader architecture. These tests, carried out by the development team, are designed to detect logical and functional errors at the module level, ensuring each function operates as expected in isolation. Unit tests were applied to both the web platform and the mobile application, enabling the early identification of issues and supporting the continuous improvement of system reliability (Siqueira & Davis, 2021).
Testing procedures were implemented in Visual Studio for the web modules and API endpoints, including validations for client registration and data transmission related to pet records (Web API Search: Discover Web API and Its Endpoint with Natural Language Queries | SpringerLink, s. f.). Results indicated an 80% pass rate, with identified coding issues promptly addressed during subsequent development cycles.
3.2 Usability testing
Usability testing was conducted to assess the overall user experience and to ensure that the system’s design aligns with the operational needs of its primary users. These tests provided valuable insights into how easily users, particularly veterinary staff and administrative personnel, interact with the platform to perform routine tasks. Feedback collected during this evaluation process allowed for the identification of interface elements requiring refinement, while also confirming that the system meets key usability criteria such as accessibility, task efficiency, and intuitive navigation.
The evaluation yielded positive outcomes across all measured dimensions. As summarized in Table 2, the average scores for usability items ranged from 3.60 to 4.50 on a 5 point Likert scale, reflecting a generally favorable user experience. The highest mean score was observed for Overall Satisfaction (M = 4.50, SD = 0.53), indicating a strong level of user acceptance and satisfaction. Similarly, Feature Completeness (M = 4.40, SD = 0.52) and Ease of Use (M = 4.30, SD = 0.67) received consistently high ratings, suggesting that users found the system both functional and user friendly.
The lowest score was noted for System Response Time (M = 3.60, SD = 0.52), which, while still within an acceptable range, highlights an area for potential optimization in future iterations. The relatively low standard deviations across all categories reflect uniform responses among participants, further supporting the system’s reliability and consistency in clinical application.
3.3 Interface testing
Interface testing focused on verifying compliance with visual and interactive design standards expected by the end user. Evaluations included reviews of layout consistency, graphical arrange-ment, element alignment, and system respon-siveness to user actions. Tests were applied to both the web and mobile interfaces, and findings highlighted several design inconsistencies that warranted refinement to enhance the overall user experience.
3.4 Functionality testing
Functionality testing was done to make sure that all the system's main features worked as they were supposed to, as outlined in the design phase. This process included running test cases for key modules in a planned way, such as the workflows for registering veterinarians and clients. The goal was to check how the system worked with certain inputs and find any functional problems. Most of the features worked as they should, but there were some problems, especially missing administrative features. These were fixed so that the system met the needs of all stakeholders.
Table 2
Usability responses summary
| Ease of use | Clarity of interface | System response time | Feature completeness | Overall satisfaction |
mean | 4.3 | 4.3 | 3.6 | 4.4 | 4.5 |
median | 4.0 | 4.0 | 4.0 | 4.0 | 4.5 |
std | 0.67 | 0.67 | 0.52 | 0.52 | 0.53 |
4. Conclusions
The Animal Place veterinary clinic met all of its goals and requirements after successfully putting the web and mobile system into place. Based on the process of developing and evaluating, we can draw the following conclusions.
It was very important to use qualitative data collection methods, especially semi-structured interviews, to understand the veterinary clinic's specific needs and how it works. These insights helped us determine the right clinical variables and design the system's features so that both the web and mobile apps could solve real-world diagnostic problems (Janke et al., 2021).
Using data science methods, especially the Knowledge Discovery in Databases (KDD) process, made it possible to organize the steps of data preprocessing, transformation, and choosing the best machine learning model. The symptomatology dataset was cleaned up and put into a CSV file, and then data mining tests were run on the Weka platform. The J48 decision tree model had the highest accuracy of all the algorithms tested, with a precision rate of 95%. This evidence proves that it is a good choice for the diagnostic task.
During the system design phase, it was crucial to use Unified Modeling Language (UML) diagrams. These diagrams made it easy to see how the system was set up, what roles users played, and how data flowed. These diagrams made it easier to create a database model that was consistent and could grow. PostgreSQL was used to create the final relational database, which had clear entities, attributes, and relationships that met the application's functional needs(Nazareno González & Quiroz García, 2025).
The Extreme Programming (XP) method was used to build the system. This made progress faster and easier and it also made it easier for the team to work together. C# and .NET Core were used to build the web app, and Java and Kotlin were used to build the Android app. These technologies worked well together to create a strong and modular system architecture(Priya & Punithavathy, 2022). An API layer was added to handle database requests and ensure that all the parts worked together smoothly. In the end, the final product is a diagnostic platform that works perfectly and can be scaled up in the future to add more modules and services.
While the developed system demonstrated satisfactory performance and fulfilled its core objectives, several limitations emerged throughout the research and implementation process that warrant consideration for future improvements.
One of the main limitations was the size and diversity of the dataset used to train the machine learning model. Although the model achieved high accuracy with the available data, the dataset was limited to symptomatological records from a single veterinary clinic, which may restrict the generalizability of the system to other clinical contexts or geographic regions. Additionally, the symptoms recorded were primarily based on practitioner input, which may introduce subjectivity and potential variability in labeling.
Another limitation lies in the scope of diseases addressed by the system. Currently, the platform focuses exclusively on the detection of hemoparasitosis. Expanding the diagnostic capabilities to include other common infectious and parasitic diseases in canines would significantly increase the system’s utility and clinical relevance.
From a technical perspective, the integration between the mobile app and the API, although functional, could benefit from the incorporation of advanced authentication mechanisms and offline capabilities to improve data security and resilience in low-connectivity environments. Furthermore, the chatbot interface, while advantageous for general inquiries, is currently limited in its ability to respond to complex clinical scenarios and lacks natural language adaptability beyond predefined intents.
Future work will focus on addressing these limitations by expanding the clinical dataset through collaborations with other veterinary institutions and enhancing the model’s training with larger and more diverse samples. Additional development will also include the implementation of multi-disease diagnostic modules, refinement of the conversational agent using advanced NLP frameworks, and incorporation of real-time data analytics to support evidence-based decision-making. These enhancements aim to transform the system into a comprehensive digital tool for veterinary diagnostics, adaptable to broader contexts and evolving clinical demands.
Acknowledgment
The authors would like to express their sincere gratitude to the staff of the Animal Place veterinary clinic for their invaluable collaboration throughout the development and testing phases of the system. Their professional insights, availability for interviews, and openness to technological innovation were essential to the success of this project.
Referencias bibliográficas
Adams, D. J., Rosenberg, D. E., & Yirui, H. (2016). Prevalence of vector-borne diseases in a sample of client-owned dogs on Santa Cruz in the Galápagos Islands: A pilot study. Veterinary Parasitology: Regional Studies and Reports, 6, 28-30. https://doi.org/10.1016/j.vprsr.2016.11.007
Aguirre-Munizaga, M., Briones-Zambrano, M., & Jurado-Chagerben, A. (2025a). Sistemas de Información Gerencial como una Herramienta Clave para la Toma de Decisiones Empresariales. MQRInvestigar, 9(1), e138. https://doi.org/10.56048/MQR20225.9.1.2025.e138
Aguirre-Munizaga, M., Carrasco, F., Aviles, F., Samaniego-Cobo, T., & Castro, C. M. (2025b). Web-Based System for the Diagnosis of Canine Diseases Using Data Mining Techniques. En Communications in Computer and Information Science (pp. 51-61). Springer Nature Switzerland. https://doi.org/10.1007/978-3-031-75702-0_5
Chua, A. P. B., Galay, R. L., Tanaka, T., & Yamazaki, W. (2020). Development of a Loop-Mediated Isothermal Amplification (LAMP) Assay Targeting the Citrate Synthase Gene for Detection of Ehrlichia canis in Dogs. Veterinary Sciences, 7(4), Article 4. https://doi.org/10.3390/vetsci7040156
da Silveira, J. A. G., Rabelo, É. M. L., & Ribeiro, M. F. B. (2011). Detection of Theileria and Babesia in brown brocket deer (Mazama gouazoubira) and marsh deer (Blastocerus dichotomus) in the State of Minas Gerais, Brazil. Veterinary Parasitology, 177(1), 61-66. https://doi.org/10.1016/j.vetpar.2010.10.044
Dai, W., & Ji, W. (2014). A MapReduce Implementation of C4.5 Decision Tree Algorithm. International Journal of Database Theory and Application, 7(1), 49-60. https://doi.org/10.14257/ijdta.2014.7.1.05
Elasan, S., & Yilmaz, O. (2025). Comprehensive Global Analysis of Future Trends in Artificial Intelligence‐Assisted Veterinary Medicine. Veterinary Medicine and Science, 11(3). https://doi.org/10.1002/vms3.70258
Fiuza Lemos Ferreira, B. (2022). Leishmaniose visceral canina como diagnóstico diferencial para hemoparasitoses transmitidas por carrapatos: Relato de caso. Pubvet, 16(04). https://doi.org/10.31533/pubvet.v16n04a1081.1-7
Górecka, W., Skalski, K., Wochnik, M., & Dąbrowski, R. (2025). The use of artificial intelligence in the educational process from the perspective of a veterinary medicine student. https://doi.org/10.13140/RG.2.2.27849.45926
Haryanto, I. D., & Saefurrahman, S. (2024). Implementasi Chatbot Kesehatan Kucing Melalui Dialogflow dan Telegram untuk Pemberian Informasi Penyakit dan Perawatan. JTIM : Jurnal Teknologi Informasi dan Multimedia, 5(4), 365-376. https://doi.org/10.35746/jtim.v5i4.484
Hernández Bustos, M. B., & Fuentes Terán, V. M. (2018). La Ley Orgánica de Bienestar Animal (LOBA) en Ecuador: Análisis jurídico. Derecho Animal. Forum of Animal Law Studies, 9(3), 108. https://doi.org/10.5565/rev/da.328
Janke, N., Coe, J. B., Bernardo, T. M., Dewey, C. E., & Stone, E. A. (2021). Pet owners’ and veterinarians’ perceptions of information exchange and clinical decision-making in companion animal practice. PLOS ONE, 16(2), e0245632. https://doi.org/10.1371/journal.pone.0245632
Karasová, M., Tóthová, C., Grelová, S., & Fialkovičová, M. (2022). The etiology, incidence, pathogenesis, diagnostics, and treatment of canine babesiosis caused by Babesia gibsoni infection. Animals, 12, 739. https://doi.org/10.3390/ani12060739
Kraleva, R. S., Kralev, V. S., Sinyagina, N., Koprinkova-Hristova, P., & Bocheva, N. (2018). Design and Analysis of a Relational Database for Behavioral Experiments Data Processing. International Journal of Online and Biomedical Engineering (iJOE), 14, Article 2. https://doi.org/10.3991/ijoe.v14i02.7988
Nazareno González, G. M., & Quiroz García, A. L. (2025). Sistema Web Y Móvil Para El Reconocimiento Del Gusano Cogollero En La Producción De Maíz (Zea Mays L.) Mediante la Técnica Visión Por Computadora. Revista internacional de Investigación y Desarrollo Global, 4(2), 45-59. https://doi.org/10.64041/riidg.v4i2.36
Niknejad, N., Ismail, W., Ghani, I., Nazari, B., Bahari, M., & Hussin, A. R. B. C. (2020). Understanding Service-Oriented Architecture (SOA): A systematic literature review and directions for further investigation. Information Systems, 91, 101491. https://doi.org/10.1016/j.is.2020.101491
Ord, R. L., & Lobo, C. A. (2015). Human Babesiosis: Pathogens, Prevalence, Diagnosis, and Treatment. Current Clinical Microbiology Reports, 2(4), 173-181. https://doi.org/10.1007/s40588-015-0025-z
Priya, N., & Punithavathy, E. (2022). A Review on Database and Transaction Models in Different Cloud Application Architectures. En S. Shakya, K.-L. Du, & W. Haoxiang (Eds.), Proceedings of Second International Conference on Sustainable Expert Systems (pp. 809-822). Springer Nature. https://doi.org/10.1007/978-981-16-7657-4_65
Qureshi, K. A., Parvez, A., Fahmy, N. A., Abdel Hady, B. H., Kumar, S., Ganguly, A., Atiya, A., Elhassan, G. O., Alfadly, S. O., Parkkila, S., & Aspatwar, A. (2023). Brucellosis: Epidemiology, pathogenesis, diagnosis and treatment–a comprehensive review. Annals of Medicine, 55(2). https://doi.org/10.1080/07853890.2023.2295398
Serin, H., & Körez, M. K. (2025). A Bibliometric Analysis of Research on Artificial Intelligence in Veterinary Medicine. Black Sea Journal of Agriculture, 8(3), 375-384. https://doi.org/10.47115/bsagriculture.1646312
Siqueira, F., & Davis, J. G. (2021). Service Computing for Industry 4.0: State of the Art, Challenges, and Research Opportunities. ACM Comput. Surv., 54(9), 188:1-188:38. https://doi.org/10.1145/3478680
Web API Search: Discover Web API and Its Endpoint with Natural Language Queries | SpringerLink. (s. f.). https://link.springer.com/chapter/10.1007/978-3-030-59618-7_7
Ybanez, R. H. D., Ybanez, A. P., Arnado, L. L. A., Belarmino, L. M. P., Malingin, K. G. F., Cabilete, P. B. C., Amores, Z. R. O., Talle, M. G., Liu, M., & Xuan, X. (2018). Detection of Ehrlichia, Anaplasma, and Babesia spp. In dogs of Cebu, Philippines. Veterinary World, 11(1), 14-19. https://doi.org/10.14202/vetworld.2018.14-19